Principles and challenges of genomewide DNA methylation analysis
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
REVIEWS
A p p l i c At i o n s o f n e x t- g e n e r At i o n s e q u e n c i n g
Principles and challenges of genome-
wide DNA methylation analysis
Peter W. Laird
Abstract | Methylation of cytosine bases in DNA provides a layer of epigenetic control in
many eukaryotes that has important implications for normal biology and disease. Therefore,
profiling DNA methylation across the genome is vital to understanding the influence of
epigenetics. There has been a revolution in DNA methylation analysis technology over the
past decade: analyses that previously were restricted to specific loci can now be performed
on a genome-scale and entire methylomes can be characterized at single-base-pair
resolution. However, there is such a diversity of DNA methylation profiling techniques that it
can be challenging to select one. This Review discusses the different approaches and their
relative merits and introduces considerations for data analysis.
Transposons
The phenotype of a cell is primarily determined by its DNA methylation information is erased by standard
Mobile DNA elements that can expression profile and its response to environmental molecular biology techniques, such as cloning in bacteria
relocate within the genome of cues. Epigenetics provides stability and diversity to the and PCR, and it is not revealed by hybridization as the
their hosts. cellular phenotype through chromatin marks that affect methyl group is located in the major groove of DNA rather
Restriction–modification
local transcriptional potential and that are preserved or than at the hydrogen bonds. Therefore, methylation-
system regenerated during cell division. Methylation of DNA dependent pretreatments of DNA were developed to
A set of enzymes found in many cytosine residues at the carbon 5 position (5meC) is a reveal the presence or absence of the methyl group at
bacteria and archaea that common epigenetic mark in many eukaryotes and is cytosine residues. The techniques based on these pre-
protects the host genome from
often found in the sequence context CpG or CpHpG treatments were initially restricted to relatively local-
genomic parasites. Restriction–
modification systems consist of
(H = A, T, C). When located at gene promoters, DNA ized regions of the genome, but many have now been
sequence-specific restriction methylation is usually a repressive mark. However, CpG scaled up to enable DNA methylation analysis on a
endonucleases, which target DNA methylation is increased in the gene bodies of genome-wide scale.
invading DNA, and associated actively transcribed genes in plants1–3 and mammals4–6. Although initial forays into broad DNA methylation
DNA methyltransferases with
similar recognition sequences,
Plant CpHpG methylation is found in non-expressed profiling were made with two-dimensional gel electro-
which protect the host transposons7. Bacteria and archaea also have 5meC, along phoresis8–13, the era of epigenomics truly took off with the
genome from the action of with N-4-methylcytosine and N-6-methyladenine, and adaptation of microarray hybridization techniques from
the endonucleases. these modified bases participate in restriction–modification the gene expression and genomic fields to the profiling of
systems and mismatch repair strand discrimination, histone modifications and DNA methylation patterns14–21.
Mismatch repair
A DNA-repair pathway that
among other roles. The current revolution in sequencing technology has
removes mismatched bases DNA methylation is laid down by dedicated DNA recently opened the door to single-base-pair resolution
and corrects the insertion or methyltransferases with highly conserved catalytic whole-genome DNA methylation analysis 22–24. Many
deletion of short stretches of motifs. In eukaryotes, usually only a subset of potential of the high-resolution genome-wide DNA methylation
(repeated) DNA.
target sequences in the genome are methylated, therefore profiling techniques were pioneered in model organisms
the distribution of methyl marks can convey epigenetic with small genomes, such as Arabidopsis thaliana22,24, but
USC Epigenome Center,
information by demarcating regions of transcriptional are now being applied to organisms with larger genomes,
University of Southern
California, Keck School of silence or transcriptional potential. The delineation including mammals6,23,25,26. These recent developments
Medicine, 1450 Biggy Street, of regional DNA methylation patterns, and broader in the rapidly evolving panoply of DNA methylation
Room G511B, Los Angeles, DNA methylation profiles, has important implications analysis techniques demand an updated synthesis of the
California 90089‑9601, USA. for understanding why certain regions of the genome main principles and an overview of the advantages and
e‑mail: plaird@usc.edu
doi:10.1038/nrg2732
can be expressed in specific developmental contexts disadvantages of the various approaches.
Published online and how epigenetic changes might enable aberrant This Review covers the general principles of DNA
2 February 2010 expression patterns and disease. methylation analysis, with a particular emphasis on
NATuRE REvIEWs | Genetics voluME 11 | MARCH 2010 | 191
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
CpG islands genome-scale DNA methylation analysis technologies. PCR reaction. This approach would require a thermosta-
In eukaryotic genomes, regions The relative merits of the different techniques are dis- ble DNA methyltransferase with very high efficiency
of several hundred base pairs cussed, and bioinformatic challenges that are unique and maintenance fidelity and a complete lack of de novo
that are not depleted of to DNA methylation analysis are outlined. Although methyltransferase activity, and so has not been realized
CpGs by 5-methylcytosine
deamination owing to them
some of the methods covered in this Review rely on to date. Therefore, almost all sequence-specific DNA
being unmethylated in the species-specific arrays, most of the principles could methylation analysis techniques rely on a methylation-
germ line. They often overlap be applied to any organism with 5meC, including bac- dependent treatment of the DNA before amplification
transcription start sites. Most teria and archaea. some of the methods described in or hybridization29–32. There are three main approaches:
definitions of CpG islands set a
this Review may be applicable to 5-hydroxymethylcy- endonuclease digestion, affinity enrichment and bisul-
minimum length (for example,
200 or 500 bp), a minimum
tosine, which was recently confirmed to be present in phite conversion. After genomic DNA has been treated
observed:expected CpG ratio mammalian cells27,28. with one of the methylation-dependent steps, various
(for example, greater than molecular biology techniques, including probe hybridi-
0.6 or 0.65) and a minimum Distribution and detection of DnA methylation zation and sequencing, can be used to reveal the loca-
GC content (for example,
50% or 55%).
As DNA samples are usually derived from a collection tion of the 5meC residues. The combination of different
of cells, which may vary in their DNA methylation types of pretreatment followed by different analytical
patterns, the distribution of 5meC in a DNA sample is steps has resulted in a plethora of techniques for deter-
complex. Measurements can be made either of the pat- mining DNA methylation patterns and profiles29,32–42. In
tern of methylated target sequences along individual the following sections I discuss techniques based on the
DNA molecules or as an average methylation level at three main approaches, and a summary is provided in
a single genomic locus across many DNA molecules29. TABLE 1. Expression profiling of cells treated with DNA
Analysis of DNA methylation is further complicated by methyltransferase inhibitors and/or histone deacetylase
the uneven distribution of methylation target sequences, inhibitors has also been used as a discovery tool for epi-
such as CpG, across the genome. As a consequence of genetically silenced genes. However, it is prone to false-
the frequent mutation of 5meC to thymine, these targets positive and false-negative results and is not considered
are depleted throughout most of the genome but are to be a reliable gauge of DNA methylation at a given
maintained in specific regions, such as CpG islands. This locus, and is therefore not discussed in this Review.
non-uniform distribution is an important considera-
tion in DNA methylation analysis, as discussed below. endonuclease digestion
As noted above, 5meC is not readily distinguished from Restriction endonucleases are such powerful tools in
unmethylated cytosine by hybridization-based methods molecular biology that their biological role in restric-
and, as DNA methyltransferases are not present during tion–modification systems in bacteria and archaea is
PCR or in biological cloning systems, DNA methyla- sometimes overlooked. Each sequence-specific restric-
tion information is erased during amplification. some tion enzyme has an accompanying DNA methyl-
investigators have suggested that it could be feasible to transferase that protects the endogenous DNA from
maintain the pattern of methylation during PCR if an the restriction defence system by methylating bases in the
appropriate DNA methyltransferase were present in the recognition site. some restriction enzymes are inhibited
Table 1 | Main principles of DnA methylation analysis
Pretreatment Analytical step
Locus-specific analysis Gel-based analysis Array-based analysis nGs-based analysis
enzyme • HpaII-PCR • Southern blot • DMH • Methyl–seq
digestion • RLGS • MCAM • MCA–seq
• MS-AP-PCR • HELP • HELP–seq
• AIMS • MethylScope • MSCC
• CHARM
• MMASS
Affinity • MeDIP-PCR • MeDIP • MeDIP–seq
enrichment • mDIP • MIRA–seq
• mCIP
• MIRA
sodium • MethyLight • Sanger BS • BiMP • RRBS
bisulphite • EpiTYPER • MSP • GoldenGate • BC–seq
• Pyrosequencing • MS-SNuPE • Infinium • BSPP
• COBRA • WGSBS
AIMS, amplification of inter-methylated sites; BC–seq, bisulphite conversion followed by capture and sequencing; BiMP, bisulphite
methylation profiling; BS, bisulphite sequencing; BSPP, bisulphite padlock probes; CHARM, comprehensive high-throughput arrays
for relative methylation; COBRA, combined bisulphite restriction analysis; DMH, differential methylation hybridization; HELP, HpaII
tiny fragment enrichment by ligation-mediated PCR; MCA, methylated CpG island amplification; MCAM, MCA with microarray
hybridization; MeDIP, mDIP and mCIP, methylated DNA immunoprecipitation; MIRA, methylated CpG island recovery assay;
MMASS, microarray-based methylation assessment of single samples; MS-AP-PCR, methylation-sensitive arbitrarily primed PCR;
MSCC, methylation-sensitive cut counting; MSP, methylation-specific PCR; MS-SNuPE, methylation-sensitive single nucleotide
primer extension; NGS, next-generation sequencing; RLGS, restriction landmark genome scanning; RRBS, reduced representation
bisulphite sequencing; –seq, followed by sequencing; WGSBS, whole-genome shotgun bisulphite sequencing.
192 | MARCH 2010 | voluME 11 www.nature.com/reviews/genetics
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
by 5meC in the sequence context CpG, so the patterns 4-bp recognition sequences (as opposed to the 6-bp
of cutting by such enzymes can provide a read-out of recognition sequences of SmaI and XmaI).
DNA methylation. An alternative approach is differential methylation
The most widely used methylation-sensitive restric- hybridization (DMH), which involves digestion of one
tion enzymes for DNA methylation studies are HpaII pool of genomic DNA with a methylation-sensitive
and SmaI, in part because they each have an isoschizomer restriction enzyme and mock digestion of another pool.
(MspI for HpaII) or neoschizomer (XmaI for SmaI) that is This produces parallel DNA pools that are amplified and
not inhibited by CpG methylation. That is not to say that labelled with different fluorescent dyes for two-colour
these isoschizomers are not methylation-sensitive. For array hybridization17,18,21,55. The relative fluorescent
example, MspI is inhibited by non-CpG methylation of signal intensities can be used to extract DNA methyla-
the first cytosine in the recognition sequence (CCGG). tion information at the corresponding loci on the array.
Therefore, MspI can be used to screen for meCCG DMH is one of several techniques that incorporate pre-
in plants43. cutting of genomic DNA with the restriction enzyme
The first locus-specific DNA methylation analyses in MseI. The recognition site of MseI is AATT, so it cuts
the late 1970s and early 1980s relied on digestion with frequently in the genome but relatively rarely within
methylation-sensitive restriction enzymes followed CpG islands56. This digestion step is often incorporated
by gel electrophoresis and hybridization on southern into protocols to deplete the sample of CpG-poor DNA
blots44–47. This method is still applicable for some locus- before further processing, such as adaptor ligation.
specific studies that require linkage of DNA methylation small genomes, such as the A. thaliana genome, do not
information across multiple kilobases, either between require an amplification step and can be directly labelled
CpGs or between a CpG and a genetic polymorphism. for hybridization43,57.
Methylation-sensitive restriction digestion followed An adaptation of DMH is to use the methylation-
by PCR across the restriction site is a very sensitive dependent endonuclease McrBC. This provides greater
technique that is still used in some applications today. sensitivity to densely methylated regions58,59 than using
However, it is extremely prone to false-positive results a methylation-sensitive enzyme. A further modification,
caused by incomplete digestion for reasons other than known as Methylscope, is to use McrBC to cut randomly
DNA methylation. sheared DNA (instead of MseI-digested DNA) 60,61.
optimization of tiling array design and data process-
Genome-scale approaches. over the past decade, several ing has resulted in improved performance for McrBC
endonuclease-dependent genome-scale DNA methylation digestion-based techniques. The optimized workflow
analysis approaches have been developed. Restriction is referred to as comprehensive high-throughput arrays
landmark genome scanning (RlGs) was the first tech- for relative methylation (CHARM)62. A further varia-
nique for broad DNA methylation profiling. Differences tion of DMH is to use a cocktail of methylation-sensitive
in methylation are detected as differences in the pattern of restriction enzymes to digest one pool of DNA and
restriction fragments generated by digestion with a methy- McrBC to digest the other pool63–65. This is referred to
lation-sensitive restriction enzyme separated by two- as microarray-based methylation assessment of single
dimensional gel electrophoresis. RlGs has been widely samples (MMAss)63. MMAss has been used to gener-
used to identify imprinted loci and sites that are methy- ate libraries for sanger sequencing 65. Another method,
lated in a cancer- or tissue-specific manner 8–13. simpler known as HpaII tiny fragment enrichment by ligation-
but less comprehensive gel electrophoretic techniques mediated PCR (HElP), uses ligation-mediated PCR for
include methylation-sensitive arbitrarily primed PCR the amplification of HpaII or MspI genomic restriction
(Ms-AP-PCR)48 and amplification of inter-methylated fragments followed by array hybridization62,66,67.
sites (AIMs)49, which rely on the differential patterns
of arbitrarily primed PCR products generated with Sequencing approaches. Restriction enzyme enrich-
methylation-sensitive or -resistant restriction enzyme- ment techniques are currently being adapted so that the
digested genomic DNA. The use of RlGs, Ms-AP-PCR read-out can be obtained by next-generation sequenc-
and AIMs is decreasing as these techniques are replaced ing techniques instead of array hybridization. sequence-
Isoschizomers by methods that are less labour-intensive and that do not based analysis is more flexible and powerful as it allows
Pairs of structurally distinct rely on gel electrophoresis. for allele-specific DNA methylation analysis, does not
restriction enzymes with the
same recognition sequence and
require an appropriately designed microarray, can cover
the same cleavage positions. Array hybridization. Many different techniques have more of the genome with less input DNA and avoids
been developed that couple enzymatic methods to hybridization artefacts, although it is still subject to
Neoschizomers array-based analysis. one of these is methylated CpG sequence library biases. Next-generation sequencing has
Pairs of structurally distinct
island amplification (MCA), which uses the differential been used to analyse the output of the HElP assay 67.
restriction enzymes with
the same recognition methylation sensitivities and cutting behaviours of SmaI sequencing-by-synthesis of libraries constructed from
sequence but with different and its neoschizomer XmaI. It is often combined with size-fractionated HpaII or MspI digests that are com-
cleavage positions. representational difference analysis (RDA) and, more pared with randomly sheared fragments68 is known as
recently, with array hybridization (when it is known as Methyl–seq; sequence-based analysis of HpaII diges-
Imprinted
A locus with monoallelic
MCAM)50–54. However, MCA provides lower resolution tion followed by the use of a flanking cut with a type-IIs
expression determined by the coverage than other enzyme-based techniques that rely restriction enzyme (MmeI) and adaptor ligation is known
parental origin of the allele. on pools of restriction enzymes or on enzymes with as methylation-sensitive cut counting (MsCC)6,41.
NATuRE REvIEWs | Genetics voluME 11 | MARCH 2010 | 193
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
Affinity enrichment have since been developed, including the quantitative
Chromatin immunoprecipitation (ChIP) followed by micro- direct sanger sequencing of PCR products100 and the
array hybridization (ChIP–chip) or next-generation highly automated application of this approach102.
sequencing (ChIP–seq) has proven to be a particularly
useful technique for genome-wide studies of histone Array hybridization. Bisulphite genomic sequencing
modifications69–74. similarly, affinity enrichment of excels at producing base-pair resolution DNA meth-
methylated regions using antibodies specific for 5meC (in ylation information, but bisulphite-based methods are
the context of denatured DNA) or using methyl-binding not easily adapted to array-hybridization techniques
proteins with affinity for methylated native genomic and so, until recently, were rarely used for genome-
DNA are proving to be particularly powerful tools for scale DNA methylation analysis. With the exception of
comprehensive profiling of DNA methylation in com- 5meC residues, bisulphite-treated DNA is comprised
plex genomes. Affinity purification of methylated DNA of three different bases instead of four. This reduced
was first demonstrated with the methyl-binding protein sequence complexity, and therefore greater sequence
MECP2 (REF. 56). Enrichment of methylated regions by redundancy, results in decreased hybridization spe-
immunoprecipitation of denatured genomic DNA with cificity. Hybridization of bisulphite-converted DNA
an antibody specific for methylated cytosine75, followed either requires dedicated arrays based on the bisulphite-
by hybridization to either a tiling array or to a feature converted genome or must allow for substantial
microarray, such as a CpG island array, is referred to as mismatches in the hybridization.
MeDIP76,77, mDIP78 or mCIP1. These techniques have The first mammalian hybridization arrays for ana-
been widely used to explore the methylomes of plant 1,2, lysing bisulphite-treated DNA required the ampli-
mouse75,79–81 and human62,76–78,82–88 cells. Confusingly, the fication of individual regions of the genome before
term MCIp has also been used to describe affinity puri- hybridization to a dedicated oligonucleotide array 103,104
fication of methylated native DNA by a recombinant and were therefore, in essence, modestly scaled-up
polypeptide containing the methyl-binding domain of locus-specific assays. smaller genomes, such as that of
human methyl-CpG-binding-domain protein 2 (MBD2) A. thaliana, can be hybridized to oligonucleotide arrays
fused to the Fc tail of human immunoglobulin G89–91. after whole-genome amplification using random tetra-
More recently, approaches have been developed that nucleotide primers. This method is referred to as bisul-
use higher affinity methyl-binding proteins, including phite methylation profiling (BiMP)105. It is worth noting
multimerized MBD1 domains92 and protein complexes that this approach relies on microarrays developed for
that contain the short isoform of MBD2 (MBD2b) and non-bisulphite-converted DNA. Therefore, as a result
MBD3l1 (the latter approach is called the methylated of mismatches caused by the conversion of unmethy-
CpG island recovery assay (MIRA)93–95). Also, ChIP lated cytosine residues, the overall hybridization signal
with an antibody specific for native MBD proteins has is low, both within and outside potential methylation
been used as an indirect measure of the distribution of target sequences. Regions with dense cytosine meth-
genomic methylcytosine75,96. ylation are least affected — as they retain more Cs —
These affinity-enrichment methods have mostly and yield a relatively strong signal105. Therefore, BiMP
been combined with array hybridization, in which the is only applicable to methylation-dense regions of
input DNA and enriched DNA are labelled with differ- small genomes.
ent fluorescent dyes. As with other array-based analyses, Illumina has adapted its GoldenGate BeadArray
methylcytosine affinity-enrichment techniques are now technology to interrogate DNA methylation in human
rapidly shifting to analysis by next-generation sequenc- genomic DNA samples. Multiplexed methylation-
ing techniques88. Affinity-based methods allow for rapid specific primer extension of bisulphite-converted DNA
and efficient genome-wide assessment of DNA meth- at up to 1,536 different CpG sites is performed using
ylation, but they do not yield information on individual primers that are specific for methylated and unmethy-
CpG dinucleotides and require substantial experimental lated sequences at each site106–108. The primers for the
or bioinformatic adjustment for varying CpG density at two different methylation states are labelled with dif-
different regions of the genome. ferent fluorescent dyes and the products are hybridized
Chromatin to bead arrays containing approximately 30 beads per
immunoprecipitation Bisulphite conversion CpG site. The current standard implementation for the
A technique that is used to
identify the location of
The discovery that treatment of denatured genomic human genome is the GoldenGate Methylation Cancer
DNA-binding proteins and DNA with sodium bisulphite chemically deaminates Panel I, which covers 1,505 CpG sites selected from
epigenetic marks in the unmethylated cytosine residues much more rapidly 807 genes, but custom arrays can also be designed109.
genome. Genomic sequences than methylated cytosines97,98 spurred a revolution in The GoldenGate assay can be performed on 96 sam-
containing the mark of interest
DNA methylation analysis in the 1990s99,100. This chem- ples simultaneously and is therefore well suited for the
are enriched by binding soluble
DNA chromatin extracts ical treatment of DNA effectively turns an epigenetic profiling of large numbers of specimens109–116. Illumina
(complexes of DNA and protein) difference into a genetic difference — unmethylated has also adapted its more comprehensive Infinium plat-
to an antibody that recognizes Cs are converted to Ts (by uracil) — thereby enabling form to DNA methylation analysis68,117,118. The Infinium
the mark. Related techniques many new DNA methylation detection and analysis platform incorporates a whole-genome amplification
— such as methylated DNA
immunoprecipitation — use
techniques29,32,34–42. Analysis of bisulphite-converted step after bisulphite conversion, which is followed
antibodies to recognize DNA DNA was initially done by sanger sequencing of cloned by fragmentation and hybridization of the sample to
modifications directly. PCR products from single loci99,101. Many enhancements methylation-specific DNA oligomers that are linked
194 | MARCH 2010 | voluME 11 www.nature.com/reviews/genetics
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
to individual bead types. Each bead type corresponds to representation than amplification with locus-specific
a specific DNA CpG site and methylation state. The primers. several groups have combined padlock capture
current implementation of the Illumina Infinium of mammalian DNA with next-generation sequenc-
assay for DNA methylation analysis (known as the ing to achieve 90–99% target specificity after pooling
HumanMethylation27 DNA Analysis BeadChip) inter- tens of thousands of probes for bisulphite-converted
rogates 27,578 CpG sites from 14,495 protein-coding DNA in a technique called bisulphite padlock probes
gene promoters and 110 microRNA gene promoters117. (BsPP)6,25,41,123. However, as capture occurs after bisul-
phite conversion, there is again the concern that the
Sequencing approaches. Although the adaptation of methylation state could influence capture efficiency and
bisulphite-converted DNA to array hybridization has therefore distort the DNA methylation measurements,
been challenging, bisulphite-converted DNA is par- even if all methylation permutations are included in
ticularly well suited for sequencing-based approaches the capture probes. Alternatively, capture probes can
and is now enjoying a resurgence thanks to the appli- be designed to avoid CpG dinucleotides in organisms
cation of next-generation sequencing platforms. ultra- in which DNA methylation is largely restricted to
deep sequencing of a limited number of loci has been CpGs. However, this design approach severely limits
achieved by direct pyrosequencing of PCR products119 the comprehensive capture of CpG-rich regions, such
and by sequencing of more than 100 PCR products in as CpG islands.
a single run at an average coverage of more than 1,600 The ultimate comprehensive single-base-pair
reads per locus on the Roche 454 platform120. resolution DNA methylation analysis technique is
The main challenge in sequencing bisulphite- whole-genome bisulphite sequencing. Whole-genome
converted DNA arises from its low sequence com- shotgun bisulphite sequencing (WGsBs) has been
plexity. Reduced representation bisulphite sequencing achieved on the Illumina Genome Analyzer platform
(RRBs) was introduced to reduce sequence redun- for small eukaryotic genomes, such as A. thaliana22,24,
dancy by selecting only some regions of the genome and for mammalian DNA23. Increased read lengths and
for sequencing by size-fractionation of DNA frag- paired-end sequencing strategies have aided the
ments after BglII digestion121 or after MspI digestion26. implementation of WGsBs124–126, although approxi-
These choices of restriction enzymes enrich for CpG- mately a tenth of the CpG dinucleotides in the mam-
containing segments of the genome but do not target malian genome remain refractory to alignment of
specific regions of interest in the genome. Targeting has bisulphite-converted reads.
been accomplished by array capture or padlock capture
before sequencing. Targeted capture on fixed arrays or Other approaches. An alternative approach to sequenc-
by solution hybrid selection can enrich for sequences tar- ing or hybridization is detection by matrix-assisted
geted by a library of DNA or RNA oligonucleotides and laser desorption ionization time-of-flight (MAlDI-
can be performed before or after bisulphite conversion. ToF) mass spectrometry 127–129. Although this approach
The advantage of capture before bisulphite conversion requires gene-specific amplification, and should there-
Array capture
is that a standard genomic array can be used and the fore be considered a candidate gene method, it is ame-
A method for enriching whole
genomic DNA for many methylation status of the region does not influence nable to automation, as implemented in the EpiTYPER
regions of interest by the capture. However, array capture is a relatively inef- platform developed by sequenom, which relies on
hybridization to an array ficient process, particularly for GC-rich target regions gene-specific amplification of bisulphite-treated DNA
containing RNA or DNA such as CpG islands, which are of particular interest in followed by in vitro transcription, base-specific RNA
sequences complementary
to the regions of interest.
DNA methylation studies. Targeted capture requires cleavage and MAlDI-ToF analysis128,129. Although it is
relatively large quantities of native genomic DNA or not a genome-wide technology, it can provide quantita-
Padlock capture amplification before hybridization, which would erase tively accurate results at multiple CpG dinucleotides for
A method for simultaneously DNA methylation information in native genomic DNA. hundreds of gene loci128,130 and can be reliably applied
capturing and amplifying large
solution hybrid selection is more efficient but allows to pooled DNA samples to obtain group averages for
numbers of regions of interest
from whole genomic DNA. complex cross-hybridization structures, resulting in valuable samples130.
Each padlock probe has two more enrichment of off-target sequences.
complementary oligonucleotide Alternatively, array capture after bisulphite conver- relative merits of different approaches
sequences that flank a region of sion and adaptor-mediated PCR amplification provides A straightforward comparison of DNA methylation
interest. The sequences are
joined by a loop of DNA that
sufficient DNA for constructing a sequencing library analysis methods is hampered by the complexity and
ensures efficient joint (referred to here as bisulphite conversion followed diversity of the various techniques. Many approaches
hybridization and contains by capture and sequencing (BC–seq))122. Although have competing strengths and weaknesses. The choice
sequences for PCR with this involves small amounts of input DNA, it requires of technique will be influenced by the number of sam-
universal primers.
multiple permutations of capture oligonucleotides to ples and the quality and quantity of DNA, as well as the
Solution hybrid selection reflect different methylation states and is subject desired coverage and resolution. Also, it is necessary to
A method for enriching whole to potential measurement error caused by varying capture take into account the organism that is being studied:
genomic DNA for many regions efficiency of different methylation states122. array-based analysis requires that a suitable array for
of interest by hybridization Padlock capture provides improved enrichment the species of interest is available, whereas sequence-
to a complex library of RNA
or DNA sequences in solution,
efficiency by combining the increased annealing spe- based analyses are generally applicable to any species for
followed by retrieval of the cificity of two tethered probes, and subsequent amplifi- which a reference genome exists. A summary of features
annealed hybrids. cation with universal primers allows for a more uniform and potential sources of bias is shown in TABLE 2.
NATuRE REvIEWs | Genetics voluME 11 | MARCH 2010 | 195
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
Table 2 | features and sources of bias for various techniques
technology Features Potential sources of bias
Unambiguous identification
Good coverage of regions
Cross-hybridization bias
measurement capability
Non-CpG methylation
amounts of input DNA
Incomplete bisulphite
with low CpG density
Compatible with low
In cis co-methylation
Bisulphite PCR bias
Full repeat-masked
Fragment size bias
genome coverage
of CpG measured
DNA methylation
CpG density bias
GC content bias
conversion bias
Allele-specific
Copy-number
variation bias
information
information
status bias
Infinium (•) • • • •
Enzyme–chip (•) (•) (•) • • •
MeDIP–chip • • • • •
BSPP • • • • • • •
BC–seq • • • • • • •
RRBS • • • • • • •
Enzyme–seq • • • (•) • •
MeDIP–seq • • • • •
WGSBS • • • • • • • • •
‘•’ indicates that the method has this feature or potentially has this bias; ‘(•)’ indicates that the method has this feature to a limited extent or in some circumstances.
BC–seq, bisulphite conversion followed by capture and sequencing; BSPP, bisulphite padlock probes; –chip, followed by microarray; MeDIP, methylated DNA
immunoprecipitation; RRBS, reduced representation bisulphite sequencing; –seq, followed by sequencing; WGSBS, whole-genome shotgun bisulphite sequencing.
Sample requirements. The many DNA methylation MeDIP and bisulphite-based techniques both use
analysis techniques differ vastly in sample requirements. ssDNA and are therefore compatible with DNA sam-
some techniques, such as RlGs, require in excess of ples that have been previously denatured. Enzyme-based
2 μg of high-purity, high-molecular-weight genomic techniques and MIRA both act on dsDNA. This has the
DNA. others, such as the Illumina GoldenGate tech- potential advantage that these methods could in princi-
nology, are compatible with degraded DNA extracted ple be applied to native DNA before purification — for
from formalin-fixed, paraffin-embedded samples131. As example, they could be used to detect tumour-derived
a general rule, methods based on methylation-sensitive methylated DNA in serum or plasma.
or methylation-dependent endonuclease treatment tend Bisulphite treatment not only requires DNA dena-
to require DNA of high purity, quantity and integrity, turation before treatment but also causes substantial
although adaptations of these techniques can reduce DNA degradation, and purification is needed to remove
sample quantity requirements67. Affinity-enrichment the sodium bisulphite. For these reasons, input DNA for
techniques are more tolerant of DNA impurity and many bisulphite-based methods can be of low purity
integrity, but many require substantial quantities of and integrity. In fact, crude lysis without subsequent
input genomic DNA to produce sufficient enriched purification can be sufficient preparation of samples for
output DNA. The ratio of input DNA to affinity reagent bisulphite-based analysis.
can affect the enrichment efficiencies of regions with PCR amplification is used in many locus-specific
varying methylcytosine density and should be carefully applications of bisulphite-treated DNA. It is common
considered when making adjustments to the amount of practice in PCR to use duTP during amplification
input DNA in affinity-enrichment methods. The fairly and to pretreat subsequent samples with uracil DNA
high DNA quantity and quality requirements of many glycosylase to remove carry-over PCR products from
enzyme-dependent and affinity-enrichment meth- prior reactions. This quality-assurance step is incom-
ods have resulted in the use of cell lines, as opposed patible with standard bisulphite conversion methods,
to primary tissues, in many early mammalian profiling as the bisulphite conversion deaminates unmethylated
studies. low quantities of input DNA can be mitigated cytosines to uracil, which would lead to degradation of
by amplification methods, such as PCR and whole- the bisulphite-converted genomic DNA by the uracil
genome amplification, but only if amplification is DNA glycosylase. Bisulphite deamination without
implemented after the methylation-dependent step, desulphonation before uracil DNA glycosylase treatment
otherwise the methylation information will be erased. can resolve this incompatibility 132,133.
For example, the Illumina Infinium platform incorpo-
rates whole-genome amplification after bisulphite con- Sample throughput. sample throughput is generally
version and as a result can profile 27,578 CpG sites using limited by the cost of labour and reagents. Highly auto-
less than 1 μg of input genomic DNA. mated techniques have low labour costs but are often
196 | MARCH 2010 | voluME 11 www.nature.com/reviews/genetics
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
Number of CpGs analysed per sample genome The coverage and resolution of enzyme-based meth-
1 10 102 103 104 105 106 107 108 ods are closely linked to the distribution of recognition
sequences throughout the genome. Techniques relying
MethyLight
104 on HpaII/MspI sites (HpaII and MspI are isoschizomers)
EpiTYPER can assess up to 98.5% of CpG islands and 91.1% of
GoldenGate NCBI Reference sequence (Refseq) promoters in the
human genome, but at modest resolution within each
Number of samples analysed
103 MSP Infinium
locus as a result of the average distance between HpaII/
COBRA Enzyme–chip MspI sites within CpG islands67. The low-specificity of
102
Pyrosequencing MeDIP–chip the recognition site for McrBC (RmeC…RmeC, in which R
Southern blot BSPP is A or G) suggests that McrBC-based analysis can reveal
BC–seq DNA methylation at almost any methylated CpG island
10 Sanger BS
in the genome, but the requirement of two methylated
Enzyme–seq
cytosines within a restricted distance from each other
RRBS in cis (40–3,000 bp) results in a nonlinear relationship
MeDIP–seq between DNA methylation density and cutting efficiency.
1
WGSBS The effective coverage and resolution of enzyme-based
array hybridization techniques is a function of the dis-
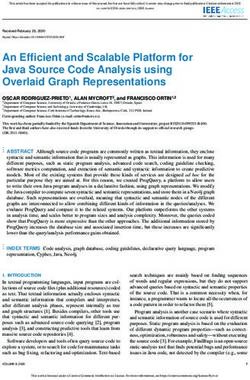
Figure 1 | sample throughput versus genome coverage. A plot of sample throughput tribution of potential cleavage sites and the composition
Nature
against genome coverage for various DNA methylation techniques. Reviews | Genetics
Throughput is of the hybridization array 62. likewise, the coverage and
determined by the number of samples that can be analysed per experiment, based on resolution of affinity methods depends on the genomic
large eukaryotic genomes. Coverage is determined by the number of CpGs in the distribution of potential affinity targets (for example,
genome that can be analysed per experiment. BC–seq, bisulphite conversion followed
CpG) and array composition. using next-generation
by capture and sequencing; BS, bisulphite sequencing; BSPP, bisulphite padlock
probes; –chip, followed by microarray; COBRA, combined bisulphite restriction
sequencing instead of microarrays to generate data
analysis; MeDIP, methylated DNA immunoprecipitation; MSP, methylation-specific output from enzyme-, affinity- and bisulphite-based
PCR; RRBS, reduced representation bisulphite sequencing; –seq, followed by methods eliminates some of the limitations that are
sequencing; WGSBS, whole-genome shotgun bisulphite sequencing. attributed to array composition. However, the base-
composition and fragment-size biases of sequencing
platforms can introduce new coverage limitations and/or
reagent-intensive. The labour involved in many of the measurement error 134.
current enzyme-based and affinity-enrichment meth- Furthermore, the sequence context of the methyl-
ods precludes the processing of large numbers of samples. cytosine residue needs to be considered when selecting
Most studies that have used array-hybridization or next- an appropriate technique. A brief summary of these
generation sequencing applications have reported results considerations is provided in BOX 1.
for just a few samples at most. By contrast, semi-automated
bisulphite-based technologies, such as the sequenom Accuracy and reproducibility. DNA methylation profil-
EpiTYPER and Illumina GoldenGate platforms, excel ing methods vary in their accuracy, reproducibility, types
at characterizing large numbers of samples, and stud- of bias and ability to detect in-cis methylation patterns, as
ies involving in excess of 100 primary human samples well as the extent to which they are influenced by target
have been reported109–111,114,115. Cost, sample through- sequence density or DNA methylation density (TABLE 2).
put and coverage are interdependent. FIG. 1 compares Methylation-sensitive restriction enzyme-based methods
throughput with genome coverage (see also below). are able to resolve methylation differences in low-CpG-
density regions, whereas McrBC-based and affinity-based
Genome coverage and resolution. The concept of reso- methods perform better for CpG-rich regions62. Many
lution is not straightforward when comparing various of the enzyme-based techniques reveal DNA methyla-
technologies with differing DNA fragmentation sizes tion information by DNA fragment length differences.
and non-uniform spacing of targeted regions. As an Fragment length can influence array hybridization
example, bisulphite sanger sequencing has been used to efficiency, but is particularly problematic in sequence-
assess approximately 40,000 different CpG dinucleotides based analysis, in which library construction efficiency
located on 2,524 different amplicons102, whereas the is strongly influenced by fragment length62,134.
Illumina Infinium DNA methylation platform has been Next-generation sequencing is often used differently
used to interrogate 27,578 different CpG dinucleotides in affinity or enzyme pretreatment experiments and
located at 14,495 different gene promoters68. As the bisulphite genomic sequencing experiments. In affin-
methylation states of CpG dinucleotides located within ity or enzyme pretreatment, short-read sequencing is
the same amplicon are highly correlated102, the number used to determine the prevalence of different regions
of independent epigenomic data points obtained on the of the genome in the enriched sample by counting the
Illumina platform is much higher (14,495 loci versus number of reads that are uniquely alignable to regions of
2,524) than in the sanger sequencing example, even the genome. By contrast, bisulphite sequencing extracts the
though the total number of CpG dinucleotides assessed alignment and DNA methylation information from
in the sanger sequencing example is slightly higher the sequence itself. The read count methods are prone
(40,000 versus 27,578). to sources of bias, such as GC content, fragment size and
NATuRE REvIEWs | Genetics voluME 11 | MARCH 2010 | 197
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
Box 1 | considerations for detection of methylation in different sequence contexts
Of the approaches for detecting DNA methylation discussed in this Review, enzyme-based methods are the most
narrowly confined by the sequence context of the 5-methylcytosine (5meC) residues. Most techniques that rely on
enzymes have been designed for either an meCpG context (in the case of restriction enzymes) or an RmeC (R = A or G)
sequence context (in the case of McrBC). Therefore, enzyme-based techniques will tend to be oblivious to meC residues
in other sequence contexts.
Most affinity-based methods will enrich for 5meC regardless of sequence context. However, the array hybridization or
sequencing used subsequently for analysis will not be able to distinguish between CpG and non-CpG methylation.
Bisulphite sequencing is the most reliable method for identifying meC in any sequence context. However, verification
following more extensive bisulphite treatment and/or recurrent observation in independent samples is recommended
to distinguish apparent CpH (H = A, T, C) methylation from incomplete bisulphite conversion.
Hydroxymethylcytosine is best detected using thin-layer chromatography, high-performance liquid chromatography
and/or mass spectrometry27,28. None of these techniques is well-suited for locus-specific analysis. Hydroxymethylcytosine
is likely to be indistinguishable from 5meC in most restriction enzyme-based techniques and in bisulphite conversion
techniques. It remains to be seen whether affinity-based methods show cross-affinity to hydroxymethylcytosine.
copy-number variations in the source DNA, that affect In sequence-based analysis, aside from flagging known
the likelihood that a particular region is included in the sNPs, the standard method for discriminating between
sequenced fragments134. The bisulphite-based methods evolutionary and bisulphite-induced deamination is to
are also subject to these effects, but they do not influ- resequence the non-bisulphite-converted experimen-
ence the DNA methylation measurement itself, as this tal genome. However, a more efficient method relies
information is extracted from the sequence. on the recognition that a sNP caused by evolutionary
Bisulphite-based methods tend to be fairly accu- deamination of meC to T will have been propagated on
rate and reproducible. The major sources of bias and the opposing DNA strand as an A, whereas bisulphite
measurement error are incomplete bisulphite conver- deamination of an unmethylated cytosine will leave the
sion and differential PCR efficiency for methylated G on the opposing strand unaffected137. sequencing of
versus unmethylated versions of the same sequence135. both DNA strands of bisulphite-converted DNA can
Incomplete bisulphite conversion can arise from incom- therefore discriminate between a CpG sNP and an
plete denaturation before bisulphite treatment or rean- unmethylated CpG without the need to resequence the
nealing during the bisulphite conversion. Bisulphite non-bisulphite converted genome. sufficient coverage
conversion destroys the self-complementarity of DNA. can resolve heterozygotes and randomly hemimethylated
Therefore, repeated denaturation cycles during the bisul- states. Interestingly, the methylation state of CpG dinu-
phite conversion process are usually sufficient to ensure cleotides can be associated with cis-linked sNPs138.
near complete conversion, as reannealing becomes pro- Therefore, sequence-based technologies that can pro-
gressively less likely. The completion of bisulphite con- vide allele-specific DNA methylation information are
version can be monitored with assays that are specifically preferable to allele-agnostic methylation assays.
designed to detect incomplete conversion136 by monitor- Enzyme-based analyses and enrichment techniques
ing the conversion of spiked DNA controls or by reten- both rely on the relative enrichment or depletion of
tion of non-target sequence cytosine dinucleotides, such regions of the genome. The occurrence of aneuploidy
as CpH in most mammalian genomes, with the caveat in cancer cells can lead to inaccuracies in methylation
that native CpH methylation may exist, particularly in measurements if copy-number alterations are not appro-
embryonic stem cells23,26,120. It should be noted that over- priately controlled. Enzyme-based methods that rely on
treatment with bisulphite not only degrades DNA but the balanced measurement of the ratio between methyl-
can also lead to an increased incidence of methylated ated and unmethylated versions of a sequence are less
cytosines converting to thymine residues98, which results prone to measurement error caused by copy-number
in under-reporting of DNA methylation. This can also alterations than methods that rely on unbalanced
be monitored by spiked methylated DNA controls. enrichment for either just methylated or just unmethyl-
ated sequences. Affinity-based methods are particularly
Allele specificity, polymorphisms and copy-number var- prone to measurement error caused by copy-number
Hemimethylated iation. CpG dinucleotides are common sites of polymor- alterations, as they are oblivious to the unmethylated ver-
Methylation of a residue phism, owing to deamination of methylated cytosines sion of a sequence. The inclusion of a control produced
on one strand within a during evolution. The deamination event can be present from the experimental sample, such as DNA artificially
palindromic target sequence
in the reference genome or in the experimental genome, methylated by M.SssI84 or digested with MspI67, can be
but not of the corresponding
residue within the palindromic and the polymorphism is indistinguishable from bisul- used to normalize copy number variations. Bisulphite-
target sequence on the phite-induced deamination of an unmethylated cytosine based methods, such as the Illumina GoldenGate plat-
complementary DNA strand. on one strand of the experimental genome. The high form, are less susceptible to copy-number variation139.
Not be confused with frequency of CpG polymorphisms can result in errors
monoallelic methylation,
in which one allele of a
in DNA methylation measurement. Indeed, polymor- Sensitivity. Genome-scale DNA methylation profiling
locus is methylated in phisms in Illumina GoldenGate BeadArray primers can techniques are not particularly well suited for the detec-
a diploid organism. masquerade as sample-specific epigenetic variation110. tion of low-frequency DNA methylation states in a DNA
198 | MARCH 2010 | voluME 11 www.nature.com/reviews/genetics
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
Table 3 | Bioinformatic resources
Resource Purpose URL Refs
Batman MeDIP DNA methylation analysis tool http://td-blade.gurdon.cam.ac.uk/software/batman 88
BDPC DNA methylation analysis platform http://biochem.jacobs-university.de/BDPC 153
BSMAP Whole-genome bisulphite sequence mapping http://code.google.com/p/bsmap 154
CpG Analyzer Windows-based program for bisulphite DNA - 155
CpGcluster CpG island identification http://bioinfo2.ugr.es/CpGcluster 156
CpGFinder Online program for CpG island identification http://linux1.softberry.com -
CpG Island Explorer Online program for CpG Island identification http://bioinfo.hku.hk/cpgieintro.html 157
CpG Island Searcher Online program for CpG Island identification http://cpgislands.usc.edu 158
CpG PatternFinder Windows-based program for bisulphite DNA - 159
CpG Promoter Large-scale promoter mapping using CpG islands http://www.cshl.edu/OTT/html/cpg_promoter.html 160
CpG ratio and GC Online program for plotting the http://mwsross.bms.ed.ac.uk/public/cgi-bin/cpg.pl -
content Plotter observed:expected ratio of CpG
CpGviewer Bisulphite DNA sequencing viewer http://dna.leeds.ac.uk/cpgviewer 161
CyMATE Bisulphite-based analysis of plant genomic DNA http://www.gmi.oeaw.ac.at/en/cymate-index/ 162
EMBOSS CpGPlot/ Online program for plotting CpG-rich regions http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html -
CpGReport/Isochore
Epigenomics Roadmap NIH Epigenomics Roadmap Initiative homepage http://nihroadmap.nih.gov/epigenomics -
Epinexus DNA methylation analysis tools http://epinexus.net/home.html -
MEDME Software package (using R) for modelling MeDIP http://espresso.med.yale.edu/medme 163
experimental data
methBLAST Similarity search program for bisulphite-modified http://medgen.ugent.be/methBLAST 164
DNA
MethDB Database for DNA methylation data http://www.methdb.de 165–168
MethPrimer Primer design for bisulphite PCR http://www.urogene.org/methprimer 169
methPrimerDB PCR primers for DNA methylation analysis http://medgen.ugent.be/methprimerdb 164
MethTools Bisulphite sequence data analysis tool http://www.methdb.de 170
MethyCancer Database Database of cancer DNA methylation data http://methycancer.psych.ac.cn 171
Methyl Primer Express Primer design for bisulphite PCR http://www.appliedbiosystems.com/ -
methylprimerexpress
Methylumi Bioconductor package for analysing DNA http://www.bioconductor.org/packages/bioc/html/ -
methylation data from Illumina platforms methylumi.html
Methylyzer Bisulphite DNA sequence visualization tool http://ubio.bioinfo.cnio.es/Methylyzer/main/index.html -
mPod Genome-wide DNA methylation viewer integrated http://www.compbio.group.cam.ac.uk/Projects/ 172
with the Ensembl genome browser p4meth.html
PubMeth Database of DNA methylation literature http://www.pubmeth.org 173
QUMA Quantification tool for methylation analysis http://quma.cdb.riken.jp 174
TCGA Data Portal Database of TCGA DNA methylation data http://cancergenome.nih.gov/dataportal -
BDPC, Bisulphite Sequencing Data Presentation and Compilation; BSMAP, Bisulphite Sequence Mapping Program; CyMATE, Cytosine Methylation Analysis Tool for
Everyone; EMBOSS, European Molecular Biology Open Software Suite; MeDIP, methylated DNA immunoprecipitation; MEDME, Modelling Experimental Data with
MeDIP Enrichment; NIH, US National Institutes of Health; QUMA, Quantification Tool For Methylation Analysis; TCGA, The Cancer Genome Atlas.
sample, as sensitivity is generally a function of sequenc- Bisulphite genomic sequencing can provide DNA
ing depth. Therefore, it is relatively expensive to achieve methylation information from individual DNA mole-
sensitive detection of low-abundance DNA methylation cules. This feature can be used to investigate cell lineages140
patterns. The introduction of next-generation sequenc- and DNA–protein interactions141. Analysis of single
ing has opened the door to efficient deep sequencing molecules was initially achieved by subcloning of PCR
of bisulphite PCR products. For example, ultra-deep products99,100 but has more recently been adapted to the
sequencing on the Roche 454 pyrosequencing platform analysis of digital PCR products by limiting dilution142–144.
can generate thousands of reads per locus, therefore This digitization of bisulphite PCR can be used to elimi-
providing the potential for the detection of rare DNA nate bias caused by the different amplification efficiencies
methylation variants119,120. of methylated and unmethylated sequences144.
NATuRE REvIEWs | Genetics voluME 11 | MARCH 2010 | 199
© 2010 Macmillan Publishers Limited. All rights reservedREVIEWS
epigenomic bioinformatics analysis (which for affinity-based methods might
The development of computational tools and resources incorporate read direction). several important statisti-
for DNA methylation analysis is accelerating rapidly 145. cal considerations that are specific to DNA methylation
TABLE 3 provides a list of some available resources. measurements are highlighted in BOX 2.
β distribution
However, the issues related to bioinformatic analysis
A continuous probability are complex and are not discussed comprehensively conclusions and future directions
distribution with an interval here. The many considerations for hybridization-based DNA methylation analysis has undergone a veritable
between 0 and 1. Two positive techniques include image and scanning artefacts, back- revolution over the past decade. We are on the verge
parameters, α and β, are used
ground correction, batch and array normalization of witnessing a deluge of sequence-based DNA meth-
to define β distributions.
and adjustments for GC content and CpG density 88. ylation data, along with other epigenomic and genomic
Median absolute deviation sequence-based analyses involve alignment to a data types. This data production will be coordinated
A measure of statistical reference genome, collapsing of clonal reads, read in national and international consortia146,147. The bot-
dispersion that is less counts or bisulphite-based analysis23, and further data tleneck in DNA methylation advances will increasingly
influenced by outliers and
extreme values than standard
deviation. It is defined as the
median of the collection of
Box 2 | An introduction to statistical issues in DnA methylation analysis
absolute deviations from the
data set’s median. DNA samples are generally derived from mixtures of cell populations with heterogeneous DNA methylation profiles.
Bisulphite sequencing-based approaches can provide a discrete DNA methylation pattern corresponding to a single
Quantile normalization
original DNA molecule. However, most techniques provide an average measurement across the sampled DNA molecules
A method for equalizing the
for a particular locus or CpG dinucleotide. For biological and historical reasons, this is usually expressed as a fraction or
total signal intensities and
distributions of probe signal percentage methylation of the total molecules assessed. For most platforms, DNA methylation measurements represent
strengths among arrays or absolute measurements for a given sample, whereas gene expression measurements are usually expressed as a
among colour channels on an differential comparison between samples. Usually, strand-specific DNA methylation or hemimethylation is not
array. It sorts all probes by considered, although monoallelic methylation — including, but not restricted to, imprinted regions — does occur in vivo.
signal strength and then The resulting measurement scale is therefore 0 to 1, or 0 to 100%, with 0 indicating that no methylated molecules were
matches probes at each rank identified and 1 or 100% indicating that all identified molecules were methylated. The fraction is calculated as M/(M + U),
position among arrays and in which M represents the signal for methylated molecules and U the signal for unmethylated molecules.
forces the values at each rank
It is important to note that this is a finite scale (β distribution) that has different statistical properties to the infinite
position to be equal. An
scale that is commonly used in gene expression array analysis. For example, β-distributed DNA methylation
identical distribution of
probe signal strengths measurements are not normally distributed and the variance of measurements within a finite scale is influenced by the
among the arrays or colour mean of the measurements; the variance of measurements with a mean near to the middle of the range can be much
channels is obtained. larger than the variance of measurements with a mean close to the limits (0 and 1). Therefore, sorting features (for
example, genes or probes) by standard deviation will result in a bias towards features with mean methylation in the
LOESS normalization middle of the range. Reducing the number of features by selecting probes with high standard deviation or median
A computationally intensive absolute deviation is a common step in unsupervised analyses of microarray data. Therefore, variance-stabilizing data
method in which a polynomial transformations or selection of probes based on a different metric should be considered. The behaviour of the distance
regression is fitted to each
metrics used to compare measurements across samples (such as fold-change, log-ratio or simple subtraction) is
point in the data and more
different for β-distributed fractions or percentages in DNA methylation measurements than for infinite ratio scales.
weight is given to data nearer
the point of interest. It is often This should be given careful consideration when selecting the most appropriate metric. Clustering and partitioning
applied to hybridization array methods used to identify subgroups of samples with similar DNA methylation profiles are being developed for
data to remove differences in β-distributed DNA methylation data115.
global signal intensity among An alternative method is to report the ratio of methylated to unmethylated molecules for a particular locus (M/U),
data sets or colour channels. usually as a log2(M/U) ratio62,152. This has gained acceptance with the increased use of microarrays in DNA methylation
analysis62, and has the advantage that many of the tools developed for gene expression data can be readily applied.
MA plot However, several points should be considered. First, many data normalization methods for gene expression data
A representation of microarray
assume that many genes are not expressed and that most genes are not differentially expressed; similar assumptions
data in which M (vertical axis)
cannot be made for DNA methylation measurements. More importantly, M and U are generally not independent, which
is the intensity ratio between
the red (R) and green (G) colour violates an assumption of many of the statistical approaches for gene expression microarrays. M and U are biologically
channels (M=log(R/G)) and A inversely correlated, but many DNA methylation platforms show them to be positively correlated if signal strength
(horizontal axis) is the mean is strongly influenced by genomic location or by probe sequence — that is, if M and U are both derived from either a
intensity (A=(logR+logG)/2). strongly or weakly hybridizing region. Furthermore, the M/U ratio may be inappropriate for situations in which the
This representation is often platform is measuring across multiple CpG dinucleotides. For example, if two CpGs are being measured and each CpG
used as a basis for normalizing is methylated at 10%, M/U will equal 0.1 if the platform assesses each CpG independently, but will equal (0.1 × 0.1)/
microarray data, with the (0.9 × 0.9) = 0.01 if the platform only registers methylation when both CpGs are methylated and the absence of
underlying assumptions that
methylation when both CpGs are unmethylated. As the number of locally grouped CpGs increases, these distortions
dye bias is dependent on signal
become quite pronounced.
intensity, that the majority of
probes do not have very An important distinction between DNA methylation measurements and gene expression measurements is that the
different signal intensities total amount of CpG methylation can differ substantially among samples. Therefore, normalization methods, such as
among channels and that quantile normalization and LOESS normalization (which is often applied to data represented on MA plots), that assume
approximately the same similar total signal across samples can remove real biological signal. It is important to note that the fluorescent dyes in
number of probes in each the Infinium DNA methylation assay do not correspond to DNA methylation states. Therefore, normalization algorithms
channel have signal intensities based on dye channel comparisons cannot be applied to Infinium DNA methylation data without modification.
that are stronger than the DNA methylation bioinformatics, biostatistics and computational biology are fertile areas of research that are under
equivalent probes in
rapid development. TABLE 3 lists several resources for DNA methylation data analysis that are currently available.
the other channel.
200 | MARCH 2010 | voluME 11 www.nature.com/reviews/genetics
© 2010 Macmillan Publishers Limited. All rights reservedYou can also read