Melanoma cultures show different susceptibility towards E1A-, E1B-19 kDa- and fiber-modified replication-competent adenoviruses
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
Gene Therapy (2006), 1–13
& 2006 Nature Publishing Group All rights reserved 0969-7128/06 $30.00
www.nature.com/gt
ORIGINAL ARTICLE
Melanoma cultures show different susceptibility
towards E1A-, E1B-19 kDa- and fiber-modified
replication-competent adenoviruses
M Schmitz1,4, C Graf1,4, T Gut1,4, D Sirena1, I Peter1, R Dummer2, UF Greber3 and S Hemmi1
1
Institute of Molecular Biology, University of Zürich, Zürich, Switzerland; 2Department of Dermatology, University Hospital, Zürich,
Switzerland and 3Institute of Zoology, University of Zürich, Zürich, Switzerland
Replicating adenovirus (Ad) vectors with tumour tissue in two of 21 melanoma cells. (3) We inserted an RGD
specificity hold great promise for treatment of cancer. We sequence into the fiber to extend viral tropism to av integrin-
have recently constructed a conditionally replicating Ad5 expressing cells, and (4) swapped the fiber with the Ad35
AdDEP-TETP inducing tumour regression in a xenograft fiber (F35) enhancing the tropism to malignant melanoma
mouse model. For further improvement of this vector, we cells expressing CD46. The RGD-fiber modification strongly
introduced four genetic modifications and analysed the viral increased cytolysis in all of the 11 CAR-low melanoma cells.
cytotoxicity in a large panel of melanoma cell lines and The F35 fiber-chimeric vector boosted the cytotoxicity in nine
patient-derived melanoma cells. (1) The antiapoptotic gene of 11 cells. Our results show that rational engineering
E1B-19 kDa (D19 mutant) was deleted increasing the additively enhances the cytolytic potential of Ad vectors, a
cytolytic activity in 18 of 21 melanoma cells. (2) Introduction prerequisite for the development of patient-customized viral
of the E1A 122–129 deletion (D24 mutant), suggested to therapies.
attenuate viral replication in cell cycle-arrested cells, did Gene Therapy advance online publication, 16 February 2006;
not abrogate this activity and increased the cytolytic activity doi:10.1038/sj.gt.3302739
Keywords: conditionally replicating adenovirus; oncolytic virus; melanoma; E1A; E1B
Introduction tumour regression in nude mice. Despite such promising
results in small animal models, clinical trials using
The use of conditionally replicating viruses including CRAds alone were so far rather disappointing unless
adenoviruses (Ads), so-called CRAds, has become an combined with standard chemotherapy or radiation
interesting option for the treatment of solid tumours.1 therapy.8
One of the approaches to achieve tissue-specific CRAds Thus, it has become evident that several features
relies on the transcriptional control of critical viral genes. of CRAds should be further improved, including for
The insertion of tissue-specific promoters upstream of example the oncolytic activity. Viral spreading through
the E1A gene, the major transactivator controlling Ad tumour tissue is not very efficient, owing to physical
replication, allowed restricted replication, for example, barriers emerging from normal connective tissues and
in prostate2 or liver cancer cells.3 Following a similar endothelial cells within tumours and/or high intra-
approach, several groups including ours recently devel- tumoral pressure.9–11 In addition, in situ tumour tissues
oped replication-competent Ads with specificity for support viral replication much less efficiently than
melanoma.4–7 This was accomplished using a combina- cultured cell lines, for example, owing to the lack of
tion of two or four copies of the mouse or human viral receptors. To accelerate cell lysis, release and cell-to-
tyrosinase enhancer element (TE) fused to the human cell spread of virus, modifications of Ad genes regulating
tyrosinase promoter (TP). In our AdDEP-TETP construct, cell death have been suggested. These include deletions
insertion of the composite TETP construct upstream of of the E1B-19 kDa gene from wt Ad (dl337), enhancing
the E1A gene was combined with a deletion of the the cytopathic effects in vitro and in vivo for a variety of
intertwined endogenous Ad enhancer/promoter (EP). different tumours.11–15 The E1B-19 kDa protein, a protein
Injection of AdDEP-TETP into xenotransplanted melano- with sequence homology to members of the Bcl-2 family,
mas but not HeLa-derived tumours led to long-lasting prevents premature death from death receptor-mediated
signalling pathways as well as from the mitochondrial
Correspondence: Dr S Hemmi, Institute of Molecular Biology pathway.16 In vivo, the E1B-19 kDa deletion not only
Zürich, University of Zürich, Winterthurerstrasse 190, CH-8057 enhanced viral oncolysis and spread in tumour tissue in
Zürich, Switzerland. immunocompetent tumour models, but the mutant Ads
E-mail: hemmi@molbio.unizh.ch
4
These authors contributed equally to this work. also were less toxic to normal tissues.14,15
Received 9 September 2005; revised 7 November 2005; accepted 7 A different rationale underlies the introduction of E1A
November 2005 mutations. Because virtually all human tumour cells,Oncolytic adenoviruses for melanoma treatment
M Schmitz et al
2
including melanoma, display aberrant E2F activity To prove loss of complex formation between our D24
resulting from cell cycle and retinoblastoma (Rb) construct and pRb, melanoma cells SK-Mel23 were
dysregulation,17,18 it was proposed that Ad mutants infected with AdDEP-TETP or AdDEP-TETP-D24 at a
lacking E2F release activity should preferentially repli- multiplicity of infection (MOI) of 20 for 24 h. Western
cate in proliferating cells.19–22 The E1A 122–129 deletion blots of cell lysates against E1A revealed the presence of
(D24), which leads to a loss of E1A binding to pRb and E1A proteins in infected cells (Figure 1a). The various
related pocket proteins without inhibiting its other E1A bands of 30–55 kDa were derived from the 9S, 10S,
transactivation functions, has been inserted in numerous 11S, 12S and 13S mRNAs.40 Immunoprecipitated E1A
CRAds, either alone19,20 or in combination with addi- from AdDEP-TETP co-precipitated Rb, in contrast to E1A
tional modifications.21–23 isolated from AdDEP-TETP-D24-infected cells, which
To overcome limited receptor expression of Ad2/5- failed to pull down Rb (Figure 1b). The reciprocal
based vectors, genetically fiber-modified viruses have experiment using the Rb antibody for immunoprecipita-
gained interest in gene therapy.24 Modifications included tion and E1A antibody for Western staining confirmed
insertion of heterologous peptides such as the RGD motif these results (not shown).
flanked on both sides by cysteine–aspartate/phenylala- To confirm the loss of the E1B-19 kDa expression
nine–cysteine residues (RGD-4C) into the fiber HI loop, cassette, melanoma SK-Mel23 cells were infected with
which allowed efficient re-directing to various av either wild-type (wt) Ad5, AdDEP-TETP, AdDEP-TETP-
integrin-expressing cells,23,25–27 including melanoma.7,28–31 D19, AdDEP-TETP-D24 or AdDEP-TETP-D24D19 at an
Alternatively, fiber swapping of the commonly used MOI of 10 or 100. E1B-19 kDa protein, E1B-55 kDa
Ad2/5 fiber with species B Ad fibers yielded an protein and different forms of E1A proteins were
extended tropism in various cell types,32–34 including determined by Western blot analysis and compared to
melanoma cells.30,31 This is due to targeting of species B the amounts of endogenous a-tubulin (Figure 1c). The
Ads to CD46, a ubiquitous cell surface receptor.35–37 Here results confirmed the loss of E1B-19 kDa expression in
we compared several rationally engineered variants of cells infected with either the AdDEP-TETP-D19 virus or
the melanoma-specific CRAd AdDEP-TETP with respect the AdDEP-TETP-D24D19 virus, unlike AdDEP-TETP-
to their cytopathic effects on a panel of melanoma cells. infected cells, but the expression of E1B-55 kDa protein
was unchanged. E1B-19 kDa expression from cells
infected with the wt Ad5 was much lower than in cells
Results infected with the TETP controlled viruses, reflecting the
tightly controlled expression of this protein by wt Ad5.41
Construction and characterization of E1A/E1B- To test if the E1B-19 kDa deletion enhances death in
and fiber-modified melanoma-specific replicating melanoma cells, M21-L4 and M980928 cells were infected
adenoviruses with AdDEP-TETP or AdDEP-TETP-D19 for 24, 40 and
Two types of modifications relating to E1A and E1B 65 h followed by cell viability assays using annexin/
genes were introduced into the melanoma-specific propidium iodide staining (Figure 1d). Annexin-V
replicating AdDEP-TETP vector described earlier.6 A detects phosphatidylserine on the outer leaflet of the
first modification consisted of a deletion of eight amino plasma membrane, typical of the early stages of
acids at position 122–129 within the E1A protein, which apoptosis, whereas propidium iodide stains dead cells.
abolishes E1A interaction with the Rb protein, thereby In the primary melanoma cell culture M980928, the
enhancing the viral replication preference in dividing but AdDEP-TETP-D19 virus led to a strongly enhanced
not cell cycle-arrested cells.19,20 The second E1 alteration appearance of dead cells at 40 h post infection (p.i.) (42
consisted of a deletion of the E1B-19 kDa gene,38 which versus 16% dead cells from the 19 kDa-expressing virus)
was expected to substantially enhance the oncolytic viral and with both cell types at 65 h p.i. (79 versus 21%, and
activity.13 This deletion of 146 bp leads to a loss of the 61 versus 4%, respectively). The appearance of distinct
open reading frame encoding the E1B-19 kDa gene but FITC-annexin staining indicative of ongoing apoptosis
not of the E1B-55 kDa gene. It was introduced by a was observed in M980928 cells at 24 and 40 h (8 versus
SacI–BstEII deletion into the E1B gene38 (for details see 3%, and 22 versus 2%, respectively), but not in the M21-
Materials and methods in Supplementary information). L4 cells. This difference may be due to cell type-specific
Thus, in a first step, the three new viruses, AdDEP-TETP- kinetics of infection or a preference of apoptosis and
D24, AdDEP-TETP-D19 and AdDEP-TETP-D24D19, were necrosis, respectively.14,42 Note also that for the primary
generated containing the individual or the combination melanoma cell culture M980928, a relative high percen-
of both mutations. In a second step, two types of tage of presumably spontaneous cell death was seen.
modifications of the fiber capsid were introduced. A Together, the data indicated that our new melanoma-
first fiber modification consisted of the introduction of specific CRAds derived from AdDEP-TETP were E1-
the RGD-4C motif into the fiber HI loop, previously modified as designed.
shown to result in efficient transduction of various CAR-
low but av integrin-positive target cells, including
melanoma cells.7,28–31 To this end, the three vectors Analysis of cytolytic efficacies of E1-modified
AdDEP-TETP-RGD, AdDEP-TETP-D24-RGD and Ad- melanoma-specific Ad vectors in CAR-positive
DEP-TETP-D24D19-RGD were constructed. Finally, we non-melanoma and melanoma cells
generated the vector AdDEP-TETP-D24D19-F35, a fiber- We compared the ability of the four melanoma-specific
swapped Ad vector with binding specificity for the Ad vectors AdDEP-TETP, AdDEP-TETP-D19, AdDEP-
species B Ad receptor CD46.36,39 Note that this virus TETP-D24 and AdDEP-TETP-D24D19 to specifically
contains the complete E3 region. The titres of all viruses induce cytopathic effects in a panel of human tumour
were determined and are summarized in Table 2. cells comprising non-melanoma 911, HeLa (cervical
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
3
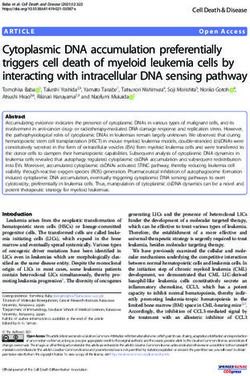
Figure 1 Molecular characterization of the E1A and E1B-19K mutated CRAds. (a) Detection of E1A proteins by Western blotting. SK-Mel23
melanoma cells were infected with the indicated viruses at MOIs of 20 and whole-cell lysates were collected 24 h p.i. The levels of E1A
proteins were determined by Western blotting using the M73 monoclonal antibody. (b) Immunoprecipitation of E1A/pRb. Total lysates from
SK-Mel23 cells infected as above were immunoprecipitated using the M73 anti-E1A antibody and protein-G-Sepharose. (c) Immunoblot
analysis of E1B-19 kDa, E1B-55 kDa and E1A expression. SK-Mel23 cells were infected with the indicated viruses and harvested 48 h later.
Cell lysates were subjected to immunoblot analysis with monoclonal E1B-19 kDa and E1B-55 kDa antibodies. E1A and a-tubulin expressions
are shown for comparison. (d) Induction of increased cell death by AdDEP-TETP-D19. M980928 and M21-L4 melanoma cells were either left
untreated or were infected with AdDEP-TETP or AdDEP-TETPD19 using an MOI of 20. Cells were analysed at the indicated time points for
cell viability using annexin/propidium iodide staining. Numbers above the horizontal line indicate the percentage of dead cells and numbers
in the bottom right quadrant indicate cells undergoing apoptosis.
carcinoma), DLD, SW480 (colon carcinoma), SKOV3 human fibroblasts, which expressed low levels of CAR
(ovary carcinoma), primary human fibroblasts, six (Table 1). The transduction efficiencies of these cells
different melanoma cell lines M21-L4, SK-Mel23, SK- were assessed with AdCMV-eGFP (results summarized
Mel25, SK-Mel28, MeWo and UKRV and the five primary in Table 1). Their susceptibility to the three new E1-
human short-term melanoma cell cultures M000301, modified AdDEP-TETP vectors was determined in
M980928, M951004, M981201 and M990802 (Table 1).43 cytopathicity assays compared to wt Ad5 and AdCMV-
All of the cells used in this experiment expressed eGFP, an E1-deleted replication-deficient virus, and the
detectable levels of the Ad primary receptor CAR, except parental strain of AdDEP-TETP. Notably, all viruses
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
4
except wt Ad5 lacked the E3 region. Figures 2a–c show E1A/B-expressing 911 cells (Figure 2a and d). To induce
the raw data and Figures 2d–f the ratios of MOI50 (MOI cytopathic effects in four non-melanocytic tumour cells
inducing 50% cell death) of wt Ad5 to recombinant (HeLa, DLD, SW480 and SKOV3), approximately 50–
viruses. Values o1 indicate reduced efficacy of the 1000 times more of the melanoma-specific viruses were
recombinant virus compared to wt Ad5 and values 41 required than wt Ad5. Both E1B-19 kDa deleted viruses
indicate higher efficiency of recombinant viruses. All five were about 20- and fivefold more cytolytic than the
recombinant viruses AdCMV-eGFP, AdDEP-TETP, Ad- parental AdDEP-TETP in HeLa and SKOV3 cells,
DEP-TETP-D19, AdDEP-TETP-D24 and AdDEP-TETP- respectively. For DLD and SW480 cells, a much smaller
D24D19 were similarly efficient as wt Ad5 in detaching increase of CPE was noticed with these viruses. The E1A
Table 1 Non-melanoma and melanoma cell cultures and summary of Ad transduction efficiency, expression of CAR and relative tyrosinase
transcript levels
Cells Tumour Transd. CARc CD46 avb5 Tyrosinase
stagea eff. (eGFP)b transcripts
(ng/0.1 ng GAPDH)d
911 (human embryonic retina cell line) ++ ND ND ND
A549 (lung carcinoma) + ++ ++++ +++ ND
HeLa (cervix carcinoma) + ++ ++++ ND 2.5773.05 104
DLD-1 (colon carcinoma) + ++ ND ND 1.8672.20 104
SW480 (colon carcinoma) + ++ ND ND 1.2571.47 104
SKOV3 (ovary carcinoma) + + ND ND 1.6872.13 104
Fibroblasts (primary foreskin fibroblasts) ND ND 2.2872.87 104
HUVEC (primary human umbilical cord vessel endothelial cells) + + ND ND ND
M21-L4 (melanoma cell line) + + ND ND 3.9472.58
SK-Mel23 (melanoma cell line) + /+ ++++ +++ 0.55570.146
SK-Mel25 (melanoma cell line) + + ND ND 1.3971.49 104
SK-Mel28 (melanoma cell line) + + ND ND 5.2371.72 102
MeWo (melanoma cell line) + + ND ND 1.7070.573 102
UKRV-Mel2 (melanoma cell line) + + ND ND 0.49370.302 102
M000301 (primary short-term melanoma) III + + ND ND 1.9370.985
M980928 (primary short-term melanoma) III + + ND ND 1.9170.828
M951004 (primary short-term melanoma) III + + ND ND 4.2272.13
M981201 (primary short-term melanoma) II + ++ ND ND 7.2674.39
M990802 (primary short-term melanoma) II + + ND ND 1.0270.39
M950322 (primary short-term melanoma) III +++ + ND
M960618 (primary short-term melanoma) III ++++ +++ ND
M960819 (primary short-term melanoma) III ++++ + ND
M980409 (primary short-term melanoma) III ++++ ++ ND
M980513 (primary short-term melanoma) III /+ ++ ++ ND
M950504 (primary short-term melanoma) I +++ + ND
M961121 (primary short-term melanoma) I ++ + ND
M991121 (primary short-term melanoma) III ND ND ND
M950710 (primary short-term melanoma) II ND
M961205 (primary short-term melanoma) I + + ND
a
Origin of the melanoma cultures: I, primary tumour; II, locoregional lymph node metastasis; III, distant metastasis.
b
Transduction efficiency determined by analysis of Ad5-based AdCMV-eGFP-mediated transgene expression; +: MOI of 100 results in mean
values 450; : MOI of 100 results in mean values o1.
c
Expression level determined by cytofluorometric analysis using the monoclonal antibody specific for CAR; assignment of expression levels:
++++: at least 16-fold shift in mean fluorescence; +++: at least eightfold shift in mean fluorescence; ++: at least fourfold shift in mean
fluorescence; +: at least twofold shift in mean fluorescence; /+: weak, but still above background shift in mean fluorescence; : negative
when compared to isotype control.
d
Determined using quantitative RT-PCR as described in Materials and methods.
ND: not done.
Figure 2 Cytolytic activity of CRAds for non-melanoma and melanoma cells. A total of 104 cells were seeded into 96-well plates and viruses
were added in 10-fold dilutions, in the range of MOI of 100–0.001. When MOI 0.1 of wt Ad5 killed approximately 50% of the cells, the
remaining cells were fixed with methanol and stained with crystal violet. (a) Non-melanoma cells included 911 helper cells expressing Ad E1
proteins, HeLa (cervical carcinoma), DLD, SW480 (colon carcinoma), SKOV3 (ovary carcinoma) and primary fibroblasts. (b) Melanoma cell
lines included M21-L4, SK-Mel23, SK-Mel25, SK-Mel28, MeWo and UKRV. (c) Primary melanoma short-term cultures included M000301,
M980928, M951004, M981201 and M990802. (d–f) Cytolytic activity of different Ads for non-melanoma and melanoma cells (calculation of
cytopathic potency index of experiment (a–c)). Cell density was determined by measuring the OD570 in a microtitre plate reader taking non-
infected cells as the 100% reference value. The MOIs inducing 50% cell loss were calculated for each virus and the results plotted as the ratios
of MOI50 of wt to recombinant virus. Note that partially resistant cells like fibroblasts and SK-Mel25 could not be included in the calculation
of the cytopathic potency index. Mean values and standard deviations of triplicates from one representative experiment are shown.
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
5
modification (AdDEP-TETP-D24) did not attenuate the levels of CAR were not included in the calculation of the
cytopathicity, when compared to parental AdDEP-TETP. cytopathic potency index, as they were susceptible to
Note that the normal human fibroblasts expressing low only the highest doses of wt Ad5 virus.
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
6
A different picture emerged for the tyrosinase-positive infection with wt Ad5, AdDEP-TETP and AdDEP-TETP-
melanoma cells. Here all the four recombinant CRAds D19 (Figure 3b). The M21-L4 cell contained about 10-fold
exerted similar or even increased cytolytic effects less AdDEP-TETP-D19 virus than wt Ad5 in both the cell-
compared to wt Ad5. Melanoma cells SK-Mel25 that associated and extracellular fractions, and intermediate
expressed similarly low levels of tyrosinase as non- levels of AdDEP-TETP. Approximately 10-fold reduced
melanoma cells (Table 1) were resistant to the CRAds levels of E1B-19 kDa-deleted viruses were generated also
(Figure 2b). Strikingly, compared to the original melano- in 911 helper cells compared to wt Ad5 (Table 2).
ma-specific virus AdDEP-TETP, the E1A- and E1B- However, the reduction of virus yield was much less
deleted virus had a clear 1–2 log increased cytolytic pronounced in the M980928 cells, suggesting that there
effect on two of five melanoma cell lines (M21-L4, SK- are cell-specific features affecting virus production rather
Mel23) as well as on four of five of the primary short time than a general replication impairment of E1B-19 kDa-
melanoma cultures (M000301, M980928, M981201 and deleted viruses. In summary, the E1B-19 kDa deletion
M990802) (Figure 2b, c, e and f). A moderate increase of resulted in enhanced cytolysis, with a moderate loss of
CPE was found for melanoma cell lines SK-Mel28 and viral production.
MeWo. Of note, on one of five melanoma cell lines
(UKRV-Mel2) and for one of the five primary short-term
cultures (M951004), the E1A-modified virus AdDEP- The cytolytic efficacy of E1- and fiber-modified
TETP-D24, but not the double-modified virus AdDEP- melanoma-specific Ad vectors on CAR-negative
TETP-D24D19, was about 100-fold more efficient than melanoma and non-melanoma cells
the E1B-19 kDa-deleted mutant viruses and also more We next compared the cytolytic efficacy of the melano-
efficient than the parental AdDEP-TETP virus. The ma-specific AdDEP-TETP, AdDEP-TETP-D24 and Ad-
cytolytic efficiency of the replication-deficient AdCMV- DEP-TETP-D24D19 with equivalent viruses carrying an
eGFP was 2–4 orders of magnitude lower than wt Ad5 in RGD insertion in their fiber genes or a virus carrying the
all cell types, except helper 911 cells, suggesting that the fiber of Ad35 (AdDEP-TETP-D24D19-F35).
cytopathicity is due to viral replication. Taken together, For this, we used the non-melanoma 911, HeLa and
the E1A deletion in AdDEP-TETP did not result in a the melanoma cell line SK-Mel23, in addition to non-
deleterious effect on the cytolytic activity when tested in melanoma A549 (lung carcinoma), and 10 different,
CAR-positive melanoma cells, and the introduction of mostly CAR-negative, primary human short-term mela-
the E1B-19 kDa deletion in general increased the cyto- nomas (Table 1). Most of the cells included in this
lytic effects. experiment were characterized previously for their sur-
face expression of CD46, CD80/86 and nine different
integrins.29 None of the cells expressed detectable levels
Replication of the E1-modified melanoma-specific of CD80/86 (not shown). CD80/86 had been postulated
Ad vectors in CAR-positive melanoma and to be a receptor of species B Ads.44 Expression of the
non-melanoma cells integrins was cell type dependent. Table 1 indicates the
To assess if the cytolytic effects are due to viral expression levels of avb5, the main target of RGD fiber-
replication, yields of the four different AdDEP-TETP modified Ad.29
viruses were determined in non-melanoma and melano- All the recombinant viruses carrying fiber modifica-
ma cells. Melanoma (MeWo, SK-Mel23, M000301, tions were similarly efficient as wt Ad5 and wt Ad35 in
M990802), non-melanoma (HeLa), primary human en- 911 helper cells. Figure 4 shows the cell detachment assay
dothelial cells (HUVECs) and foreskin fibroblasts were in microtitre wells, and Table 3 gives an overview of the
infected with MOI 1. Samples were collected at different cytolytic efficiency of the recombinant viruses compared
time points (up to 5 days p.i.) and plaque-titred on 911 to wt Ad5. In addition, the melanocyte specificity of
helper cells (Figure 3a). In both non-melanoma cell types, these vectors was preserved, as 50–5000 times more
titres of the AdDEP-TETP viruses were relatively melanoma-specific viruses were required to induce
uniformly reduced by factors of 100–1000 compared to cytopathic effects in the non-melanocytic cell lines A549
wt A5, in particular at the latest time point. This and HeLa than wt Ad5 (Figure 4a). The RGD-modified
difference was more pronounced in primary cells than melanoma-oncolytic viruses AdDEP-TETP-RGD and
in HeLa (about 1 log difference). In three of four AdDEP-TETP-D24D19-RGD were about 10-fold more
melanoma cells tested (SK-Mel23, M000301, M990802), cytolytic for the non-melanoma cells A549 and HeLa,
the titres of the AdDEP-TETP viruses were within 1 log compared to their parental viruses AdDEP-TETP
range and the growth curves were similar compared to and AdDEP-TETP-D24D19, reflecting their additional
wt Ad5. MeWo cells gave somewhat reduced yields of receptor binding features.29 Somewhat surprisingly, wt
the melanoma-oncolytic viruses, which correlated with Ad35 was 13- and 1300-fold less cytolytic than wt Ad5
the intermediate expression levels of tyrosinase in these for A549 and HeLa cells, suggesting the presence of
cells and thus most likely reflects the efficacy of the Ad35-specific host restrictions in these epithelial cell
tyrosinase promoter driving viral E1A expression (Table lines. As expected, the E1/E3-deleted AdCMV-eGFP and
1). Note that in HUVEC, the E1B-19 kDa mutant viruses AdCMV-eGFP-RGD viruses had 4 log lower cytolytic
produced lower yields than the parental AdDEP-TETP effects than wt Ad5 in all CAR-positive cell types, except
and the AdDEP-TETP-D24 virus. As expected, AdCMV- 911 helper cells. For the mostly CAR-negative melanoma
eGFP did not grow in any of these cells. To test if the lack cultures, the difference between wt Ad5 and the fiber-
of E1B-19 kDa and the concomitant induction of apop- modified E1/E3-deleted vectors was reduced to 1–2 logs.
tosis affected the overall yields of virus production, we In particular, M950322, M960819 and M950710 were
quantitated the virus particles in M980928 and M21-L4 similarly sensitive towards wt Ad5 and the E1/E3-
melanoma cells and the extracellular medium upon deleted viruses. Furthermore, the E1-deleted, but
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
7
Figure 3 Growth of wt Ad5 and CRAds in different human cells. (a) Melanoma cells MeWo, SK-Mel23, M000301 and M990801 and non-
melanoma cells HeLa, primary HUVECs and fibroblasts were infected at an MOI of 1. Cells and supernatants were harvested and pooled 2,
24, 48, 72 and 120 h p.i. and virus titres determined after three cycles of freeze–thaw by plaque assays in 911 cells. Mean values of duplicates
are shown. (b) Melanoma cells M980928 and M21-L4 were infected as for (a), but cell pellets (p) and cell supernatants (s) were processed and
titrated separately.
E3-positive AdCMV-eGFP-F35 virus showed an in- According to their susceptibility, the cells were
creased cytolytic efficiency for primary melanomas grouped into three categories. A first group of cells
compared to the E1/E3-deleted viruses. including five of the 11 melanoma cell cultures (M950322,
The cytolytic susceptibility of the different mostly M960819, M980409, M980513 and melanoma cell line
CAR-low melanoma cell cultures to fiber-unmodified SK-Mel23) were highly susceptible to RGD-modified
melanoma-oncolytic CRAds varied substantially CRAd infection, with cytolytic effects at MOIp0.1
compared to the fiber-modified viruses. The fiber- (Figure 4b). Of outstanding efficiency was AdDEP-
unmodified AdDEP-TETP and AdDEP-TETP-D24 TETP-D24D19-RGD, reaching 10- to 1000-fold increased
revealed similar or moderately weaker cytolytic efficiency than the fiber-native viruses (Figure 4b and
activity compared to wt Ad5, whereas the AdDEP- Table 3). In M980409 cells, AdDEP-TETP-D24-RGD and
TETP-D24D19 was clearly more cytolytic than the AdDEP-TETP-RGD were similarly efficient as AdDEP-
E1B-native vectors, comparable to wt Ad5. The RGD TETP-D24D19-RGD, indicating that the removal of the
fiber-modified viruses were all clearly more cytolytic apoptosis inhibitor E1B-19 kDa in these cells had no
than wt Ad5. effect on cytolysis, similar to the cytolytic activity of the
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
8
Table 2 Ads used in this study
Virus name E1A promotera E1Ab E1B-19 kDa E3 Fiberc Replication PFU/ml VP/ml PFU/VP Reference
Wt Ad5 WT WT WT WT WT (Ad5) Competent 1 1011 6 1011 1/6 6
Wt Ad35 WT WT WT WT WT (Ad35) Competent 2 109 1.8 1011 1/94 ATCC
AdCMV-eGFP WT del del del WT (Ad5) Defective 2.6 109 1.9 1011 1/73 29
AdCMV-eGFP-RGD WT del del del Ad5 HI-RGD Defective 3.4 109 2.8 1011 1/83 29
AdCMV-eGFP-F35 WT del del WT WT (Ad35) Defective 5.2 109 9.4 1011 1/181 34
AdDEP-TETP del/TETP WT WT del WT (Ad5) Competent 1.4 1010 5.8 1011 1/41 6
AdDEP-TETP-RGD del/TETP WT WT del Ad5 HI-RGD Competent 1.2 108 1.4 1010 1/117 This study
AdDEP-TETP-D24 del/TETP D24 WT del WT (Ad5) Competent 5.7 109 2.1 1011 1/37 This study
AdDEP-TETP-D24-RGD del/TETP D24 WT del Ad5 HI-RGD Competent 2.2 109 1.0 1011 1/47 This study
AdDEP-TETP-D19 del/TETP WT del del WT (Ad5) Competent 3.4 108 2.1 1010 1/62 This study
AdDEP-TETP-D24D19 del/TETP D24 del del WT (Ad5) Competent 6.8 108 6.8 108 1/64 This study
AdDEP-TETP-D24D19-RGD del/TETP D24 del del Ad5 HI-RGD Competent 1 108 2.6 1010 1/260 This study
AdDEP-TETP-D24D19-F35 del/TETP D24 del WT WT (Ad35) Competent 9 108 5.9 1010 1/66 This study
a
Wt or deleted and replaced by tyrosinase enhancer/tyrosinase promoter (TETP).
b
D24: deletion of amino acid 122–129 sequence in E1A.
c
Wt or insertion of integrin-binding arginine–glycine–aspartate (RGD) motif flanked on both sides by cysteine–aspartate/phenylalanine–
cysteine residues into fiber HI loop.
untargeted viruses. Intriguingly, the AdDEP-TETP- lytic Ads towards improving cell-to-cell spread and lytic
D24D19-F35 virus was about was 5- to 100-fold less potency are leading to more elaborate conditionally
efficient than the corresponding RGD-modified CRAd in replicating Ads. Here, we introduced four distinct
four of five cultures (M950322, M960819, M980513 and genetic modifications into our melanoma-specific CRAd
SK-Mel23), suggesting that avb5 integrin targeting might vector, AdDEP-TETP.6 Deletion of the antiapoptotic viral
increase cytolysis. A second category of cells including protein E1B-19 kDa enhanced the onset of apoptosis and
M960618, M961121 and M991121 was intermediately most efficiently increased the oncolytic activity in
susceptible to fiber-modified viruses at an MOI of about melanoma cells. Twelve of 14 CAR-proficient cell
1–10 (Figure 4c and Table 3). Note that all three cell types cultures, including eight of 10 melanoma cell cultures,
needed long incubation periods from 10 to 21 days to revealed higher susceptibility to E1B-19 kDa-deleted
show cytopathic effects. In M960618 and M991121 cells, CRAds, when compared to the E1B-unmodified vector.
the most efficient virus was again AdDEP-TETP-D24D19- This increase of the cytolytic potency correlates with the
RGD, whereas in M961121, AdDEP-TETP-RGD and earlier finding that an E1B-19 kDa deletion leads to
AdDEP-TETP-D24D19-RGD were comparably efficient. enhanced cell death caused by accelerated viral cell-to-
In all three cultures, the D19 mutation was more efficient cell spread.13 In several other reports, the E1B-19 kDa
than the isogenic virus containing E1B-19 kDa. The third deletion has been more effective at tumour cell killing
category of cells was weakly susceptible and required compared to wt virus.11,14,15,45 To our knowledge, we here
MOIs of 10–100 of the RGD fiber-modified viruses to present the first report, which quantitatively assesses the
reveal cytopathic effects (Figure 4d and Table 3). This oncolytic potential of a CRAd lacking the E1B-19 kDa.
group of cells included the fast-growing M950710 cells The enhanced cytolytic activity of E1B-19 kDa-deleted
and the slow-growing M950504 and M961205 cells. viruses was not restricted to melanoma cells, consistent
Again, the AdDEP-TETP-D24D19-RGD virus was the with other reports.7 It is noteworthy however that
most efficient CPE inducer among the fiber-modified despite a general increase of cytolytic activity, the CRAd
viruses. In M950504 cells, however, wt Ad35 was containing the D19 modification still exerted ample cell
more efficient than AdDEP-TETP-D24D19-RGD. Taken type specificity.
together, the deletion of E1B-19 kDa killed 10 of 11 A good replication and burst size are key features of
melanoma cultures more efficiently than the E1B-19 kDa- an effective oncolytic vector with significant potential
carrying viruses, indicating a clear cytolytic improve- in vivo. They critically depend on the time available for
ment. Although the Ad35 fiber-bearing viruses were virus assembly and egress. Importantly, our CRAd
more efficient than their fiber-native relatives, the RGD AdDEP-TETP-D19 grew to similar titres in CAR-expres-
modification resulted in a more robust enhancement of sing melanoma cells as E1B-19 kDa-containing isogenic
cytolytic activity in CAR-low melanoma cells, superior to strains, although the yields were moderately lower in
Ad35 fiber or unmodified CRAds, despite considerable some cell types, such as HUVEC cells, consistent with
variation among cell lines and patient-derived primary lower yields of E1B-19 kDa-deleted dl250 in normal
melanoma cultures. human bronchial epithelial cells.7 This contrasts with
some earlier reports suggesting that the titres of E1B-
19 kDa-deleted viruses can be severely reduced,46 but
are consistent with reports showing mildly re-
Discussion duced13,14,38,47,48 or even increased viral production.7,13,49
Clinical trials have shown the safety of tissue-specific It is possible that the differential expression of proteins
oncolytic Ads.1 The efficacy of adenoviral vectors and all involved in apoptosis protection in cancer cells affects
other viral vectors in clinical settings is still low (p10%), the potency and replication efficiency of the E1B-19 kDa
but combinatorial approaches with other therapeutics are mutants.7 The titres of all our melanoma directed CRAds
promising.8 In addition, genetic alterations of existing controlled by the tyrosinase enhancer/promoter were
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
9
Figure 4 Cytolytic activity of fiber-unmodified and modified CRAds for non-melanoma and melanoma cells. (a) Tested cells included non-
melanoma 911, A549 (lung carcinoma) and HeLa (cervical carcinoma) cells. (b–d) Tested cells included the indicated melanoma cells that
were categorized according to their sensitivity into highly (b), intermediately (c) and weakly susceptible (d) cells. Cytolytic activity was
analysed as described in Figure 2.
somewhat lower than wt Ad5. This correlates with the contributes to efficient lysis and ADP deletion prolongs
variable expression levels of tyrosinase in different cell viability.50 Accordingly, Ad variants with or without
melanoma cells (Table 1) (see Peter et al.6 and references normal levels of ADP were reported to have about
therein). Levels of tyrosinase transcripts also correlated 10-fold differences in virus yields and 10- to 100-fold
with the cytolytic activity of our CRAds. For example, all differences in CPE.21,51
non-melanoma cells and three melanoma cells with low In addition to the deletion of E1B-19 kDa, we have
tyrosinase SK-Mel25, M950822 and M9603069 did not introduced a second mutation into the E1 region of our
yield efficient CPE (latter two not shown). In contrast, CRAd, a deletion in the E1A immediate-early transacti-
intermediate and high expression levels of tyrosinase vator, which renders the protein incapable of inactivating
correlated with efficient cell killing. Another factor likely pRb. The earlier D24 mutant Addl922–947 was reported
contributing to lower virus yields is the deletion of the to be restricted in non-dividing cells, possibly due to
E3 region and thus the lack of ADP expression. ADP increased transactivation activity of E1A owing to loss of
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
10
sequestration to pRb, abrogation of E1A autoregulation,
:
¼ below,
/+ ¼ above wt Ad5 efficiency); +: 2- to 10-fold more efficient; ++: 11- to 100-fold more efficient; +++: 101- to 1000-fold more efficient; ++++: 1001- to 10 000-fold more efficient than wt Ad5; ND: not done.
alteration in the ubiquitin pathway or changes in the pRb
phosphorylation status.19,20 We have not seen any severe
attenuation effects of our D24 CRAd in normal HUVEC
The MOIs inducing 50% cell loss were calculated for each virus and the results plotted as the ratios of MOI50 of wt to recombinant virus. Assignment of cytolytic efficiency compared to wt Ad5:
and fibroblasts, possibly owing to limited growth arrests
: 1.9-fold less to 1.9-fold more efficient (
of these cells. As expected, our E1 double mutant CRAd
AdDEP-TETP-D24D19 had, however, about the same
oncolytic efficacy in cultured cancer cells as the isogenic
parent virus carrying an intact E1A gene, consistent with
reports on D24-modified melanoma-specific viruses by
others.4,52 Surprisingly, in one of five melanoma cell lines
(UKRV-Mel2) and one of the five CAR-positive primary
short-term cultures (M951004), the E1A-modified virus
AdDEP-TETP-D24, but not the double-modified virus
AdDEP-TETP-D24D19, was about 100-fold more efficient
than AdDEP-TETP-D19, and also more efficient than the
parental AdDEP-TETP virus. Overall, we expect that the
E1A mutation may render our CRAd less virulent in
non-malignant cells, resulting in a better therapeutic
index than the E1A normal virus.
: 11- to 100-fold less efficient; : 2- to 10-fold less efficient; Many cancer cells lose their ability to express CAR.53
To target CAR-negative cancer cells, we introduced two
types of fiber modifications, disulfide constrained RGD
motifs in the surface-exposed HI loop of the Ad5 fiber,
or we replaced the Ad5 fiber by the Ad35 fiber binding
to the ubiquitously expressed CD46 membrane cofactor.
The insertion of the RGD motif efficiently redirects Ad to
various av integrin-expressing cells, including melano-
ma.7,28–31 As anticipated, both the RGD- and the F35
fiber-modified viruses revealed a superior cytolytic
Table 3 Cytolytic efficiency of wt Ad35, E1-deleted Ads and melanoma CRAds compared to wt Ad5
efficiency on CAR-low cells compared to the control
viruses with native fiber indicating clear effects of
receptor targeting. Unexpectedly, our RGD fiber-mod-
ified CRAd AdDEP-TETP-D24D19-RGD was more effi-
cient than the F5/35-chimeric vector in eight of 11
melanoma cultures, and in the other three cultures, the
two viruses were equally efficient. Several comparative
transduction studies indicated that F5/3-chimeric vec-
tors performed better than RGD-modified vectors in
several cell types, but not in mouse dendritic cells or
human leukaemic cell lines.30,54–56 In a study by Rivera
: 101- to 1000-fold less efficient;
et al.31 only two of five F5/3-chimeric melanoma CRAds
performed clearly better when analysed for cytolytic
activity, whereas in the other three, the RGD-modified
vector was similar or better. Our results indicate that
CRAd targeting to integrins is more effective than to
CD46. It is unlikely to be strongly influenced by the
levels of the targeted receptors, as both av and CD46
were highly abundant on our target cells, and a high
CD46 expression, for example, in M960618, did not result
in particularly high sensitivity towards the AdDEP-
TETP-D24D19-F35 vector. Perhaps more likely is an
alternative explanation, namely that the CD46-targeted
vector carrying a capsid of Ad5 is not optimally
1001- to 10 000-fold less efficient;
constructed for infection via CD46 owing to differential
requirements of downstream receptors and signals in
Ad5 and Ad35 infections. Of particular interest here are
the av integrins, a well-characterized enhancer of natural
Ad2 and Ad5 infections.57,58 Activated integrins promote
Ad2/5 entry and intracellular transport and activate
stress responses and cell survival pathways in the
infected cells.59 We suggest that integrin targeting by
the constrained RGD motif of our CRAd may act
synergistically with the knockout of the antiapoptotic
viral regulator E1B-19 kDa. It will be interesting to
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
11
further analyse how integrin activation and signalling for 24 h at an MOI of 20. Cells were pelleted, washed
are coordinated with the regulation of cell survival with PBS and total protein lysates prepared in lysis
exerted by the E1A proteins. In the future, gene profiling buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8),
combined with systems-based analyses of the virus entry supplemented with protease inhibitors (Mini-Complete,
and exit pathways of patient-derived tumour specimens Roche, Basel, Switzerland). The lysates were sheared
may elucidate the nature underlying these findings using a G23 syringe and protein concentrations were
and further pave the way towards patient-customized determined using the BCA kit (Pierce, Rockford, IL,
oncolytic vectors. USA). For Western blot analysis, 40 mg protein of SK-
Mel23 lysates was separated by 10% SDS-PAGE and
electro-transferred to a nylon membrane. The membrane
Materials and methods was blocked by TBST (10 mM Tris pH 7.6, 150 mM NaCl,
0.1% Tween 20) plus 5% milk powder and incubated
Cell culture and viruses with monoclonal antibodies recognizing Ad5 E1B-19/
The primary human melanoma cell cultures and all other 21 kDa (DP16, Calbiochem, San Diego, CA, USA), Ad5
cells listed in Table 1 were grown as described ear- E1B-55 kDa (2A6),61 E1A (M73;62 kind gift of R Eckner,
lier.6,29,43 References for wt Ad5 (Ad5 wt300), wt Ad35, Institute of Molecular Biology, University of Zürich)
AdCMV-eGFP-F35, AdCMV-eGFP, AdCMV-eGFP-RGD or with a-tubulin (DM 1A, Sigma Switzerland, Buchs,
and AdDEP-TETP are summarized in Table 2. For Switzerland). Proteins were detected by an HRP-con-
generation of the new AdDEP-TETP vectors, see Supple- jugated anti-mouse secondary antibody (Amersham
mentary Information. Plaque-forming units (PFU) were Pharmacia, Berkhamsted, UK) and chemiluminescence
determined by end-point titration on 911 cells (expres- (Pierce, Iselin, NJ, USA). For immunoprecipitation,
sing high levels of CAR, CD46 and avb3 and avb5), and cleared cell lysates were incubated with M73 anti-E1A
concentrations of virion particles were determined antibody or anti-human Rb antibody (G3-245, BD
according to Maizel et al.60 PharMingen, San Diego, CA, USA), followed by incuba-
tion with protein-G-Sepharose (Amersham Pharmacia).
Cytopathic effect and virus growth experiments Precipitates were washed three times with lysis buffer.
Experiments to assess cytopathic potency were per-
formed in microtitre plate assays using triplicate inputs. Flow cytometry
Six serial 10-fold dilutions of the different viruses were Cytofluorometric analysis was performed using antibo-
plated in a volume of 50 ml in a 96-well dish, starting dies described previously.29,63 For detection of apoptotic/
with a concentration of 2 107 PFU/ml. All dilutions necrotic cells, cultured cells were grown in six-well
were prepared in cell culture medium. Cells were diluted plates to subconfluency and infected with different Ads
to 6.7 104 cells/ml and 150 ml of this suspension was at MOI 20. The cells were analysed for cell viability by
added per well (MOIs ranging from 102 to 104) and annexin/propidium iodide staining at 24, 40 and 65 h p.i.
observed daily. At the day when wt virus induced by collecting adherent and detached cells in PBS/20 mM
approximately 50% cell death at an MOI of 0.1, the EDTA and washing with PBS twice. Cell pellets were
medium was removed, cells fixed with methanol and resuspended in 1 binding buffer and incubated with
UV-treated to inactivate residual virus. Attached cells annexin-V-FITC and PI (BD Pharmingen) for 15 min,
were then stained with crystal violet and the dye was followed by cytofluorometric analysis (Epics XL; Coulter,
quantified at 570 nm in a microtitre plate reader. Non- Miami, FL, USA).
infected cells were included on each plate and taken as
100% surviving cells. For each virus sample, the MOI50
was calculated. Acknowledgements
To determine virus infectivity, cultured cells were
grown in 24-well plates to subconfluency. Cells were We thank L Fuchs, J Willers and P Selvam for excellent
infected at an MOI of 1 in a volume of 0.5 ml. After technical assistance, F Ochsenbein for artwork, S Rusconi
various time points (2, 24, 48, 72 and 120 h), cells were (University of Fribourg, Switzerland) for pTG-H5dl324
scraped with a rubber policeman and harvested together plasmid and M Havenga (Crucell Holland BC, The
with the infection medium. After three cycles of freeze– Netherlands) for wt Ad35 and AdCMV-eGFP-F35
thaw, cell supernatants were plaque-titred on 911 helper (rAd5Fib35 eGFP). Isabelle Peter was supported by the
cells. Alternatively, cell pellets and virus supernatants Julius Klaus Foundation, Silvio Hemmi was supported
were separated and used for titrations. Experiments were by the Cancer Society of the Kanton Zürich and the
performed in duplicates. University of Zürich and Reinhard Dummer and Urs
Greber were supported by the Swiss National Science
Real-time quantitative RT-PCR for tyrosinase Foundation (3200-063704.00/1 and 31-67002.01, respec-
tively).
transcripts
RT-PCR for determining tyrosinase transcripts in mela-
noma and non-melanoma cells was performed as
described previously.6 References
1 Kirn D, Martuza RL, Zwiebel J. Replication-selective virotherapy
Western blot and immunoprecipitation for cancer: biological principles, risk management and future
Approximately 106 SK-Mel23 cells were infected with directions. Nat Med 2001; 7: 781–787.
various viruses at an MOI of 10 and 100 and harvested 2 Rodriguez R, Schuur ER, Lim HY, Henderson GA, Simons JW,
by scraping after 48 h. Alternatively, cells were infected Henderson DR. Prostate attenuated replication competent
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
12
adenovirus (ARCA) CN706: a selective cytotoxic for prostate- 21 Doronin K, Toth K, Kuppuswamy M, Ward P, Tollefson AE,
specific antigen-positive prostate cancer cells. Cancer Res 1997; Wold WS. Tumor-specific, replication-competent adenovirus
57: 2559–2563. vectors overexpressing the adenovirus death protein. J Virol
3 Hallenbeck PL, Chang YN, Hay C, Golightly D, Stewart D, Lin J 2000; 74: 6147–6155.
et al. A novel tumor-specific replication-restricted adenoviral 22 Howe JA, Demers GW, Johnson DE, Neugebauer SE, Perry ST,
vector for gene therapy of hepatocellular carcinoma. Hum Gene Vaillancourt MT et al. Evaluation of E1-mutant adenoviruses as
Ther 1999; 10: 1721–1733. conditionally replicating agents for cancer therapy. Mol Ther
4 Nettelbeck DM, Rivera AA, Balague C, Alemany R, Curiel DT. 2000; 2: 485–495.
Novel oncolytic adenoviruses targeted to melanoma: specific 23 Cripe TP, Dunphy EJ, Holub AD, Saini A, Vasi NH, Mahller YY
viral replication and cytolysis by expression of E1A mutants et al. Fiber knob modifications overcome low, heterogeneous
from the tyrosinase enhancer/promoter. Cancer Res 2002; 62: expression of the coxsackievirus-adenovirus receptor that limits
4663–4670. adenovirus gene transfer and oncolysis for human rhabdomyo-
5 Zhang L, Akbulut H, Tang Y, Peng X, Pizzorno G, Sapi E et al. sarcoma cells. Cancer Res 2001; 61: 2953–2960.
Adenoviral vectors with E1A regulated by tumor-specific 24 Mizuguchi H, Hayakawa T. Targeted adenovirus vectors. Hum
promoters are selectively cytolytic for breast cancer and Gene Ther 2004; 15: 1034–1044.
melanoma. Mol Ther 2002; 6: 386–393. 25 Dmitriev I, Krasnykh V, Miller CR, Wang M, Kashentseva E,
6 Peter I, Graf C, Dummer R, Schaffner W, Greber UF, Hemmi S. A Mikheeva G et al. An adenovirus vector with genetically
novel attenuated replication-competent adenovirus for melano- modified fibers demonstrates expanded tropism via utilization
ma therapy. Gene Therapy 2003; 10: 530–539. of a coxsackievirus and adenovirus receptor-independent cell
7 Liu Y, Ye T, Sun D, Maynard J, Deisseroth A. Conditionally entry mechanism. J Virol 1998; 72: 9706–9713.
replication-competent adenoviral vectors with enhanced infec- 26 Koizumi N, Mizuguchi H, Hosono T, Ishii-Watabe A, Uchida E,
tivity for use in gene therapy of melanoma. Hum Gene Ther 2004; Utoguchi N et al. Efficient gene transfer by fiber-mutant
15: 637–647. adenoviral vectors containing RGD peptide. Biochim Biophys
8 Everts B, van der Poel HG. Replication-selective oncolytic Acta 2001; 1568: 13–20.
viruses in the treatment of cancer. Cancer Gene Ther 2005; 12: 27 Garcia-Castro J, Segovia JC, Garcia-Sanchez F, Lillo R, Gomez-
141–161. Navarro J, Curiel DT et al. Selective transduction of murine
9 Heise CC, Williams A, Olesch J, Kirn DH. Efficacy of a myelomonocytic leukemia cells (WEHI-3B) with regular and
replication-competent adenovirus (ONYX-015) following intra- RGD-adenoviral vectors. Mol Ther 2001; 3: 70–77.
tumoral injection: intratumoral spread and distribution effects. 28 Nakamura T, Sato K, Hamada H. Effective gene transfer to
Cancer Gene Ther 1999; 6: 499–504. human melanomas via integrin-targeted adenoviral vectors.
10 Bilbao R, Bustos M, Alzuguren P, Pajares MJ, Drozdzik M, Qian Hum Gene Ther 2002; 13: 613–626.
C et al. A blood–tumor barrier limits gene transfer to experi- 29 Nagel H, Maag S, Tassis A, Nestle FO, Greber UF, Hemmi S. The
mental liver cancer: the effect of vasoactive compounds. Gene alphavbeta5 integrin of hematopoietic and nonhematopoietic
Therapy 2000; 7: 1824–1832. cells is a transduction receptor of RGD-4C fiber-modified
11 Harrison D, Sauthoff H, Heitner S, Jagirdar J, Rom WN, Hay JG. adenoviruses. Gene Therapy 2003; 10: 1643–1653.
Wild-type adenovirus decreases tumor xenograft growth, but 30 Volk AL, Rivera AA, Kanerva A, Bauerschmitz G, Dmitriev I,
despite viral persistence complete tumor responses are rarely Nettelbeck DM et al. Enhanced adenovirus infection of melano-
achieved—deletion of the viral E1b-19-kD gene increases the ma cells by fiber-modification: incorporation of RGD peptide or
viral oncolytic effect. Hum Gene Ther 2001; 12: 1323–1332. Ad5/3 chimerism. Cancer Biol Ther 2003; 2: 511–515.
12 Barker DD, Berk AJ. Adenovirus proteins from both E1B reading 31 Rivera AA, Davydova J, Schierer S, Wang M, Krasnykh V,
frames are required for transformation of rodent cells by viral Yamamoto M et al. Combining high selectivity of replication
infection and DNA transfection. Virology 1987; 156: 107–121. with fiber chimerism for effective adenoviral oncolysis of CAR-
13 Sauthoff H, Heitner S, Rom WN, Hay JG. Deletion of the negative melanoma cells. Gene Therapy 2004; 11: 1694–1702.
adenoviral E1b-19kD gene enhances tumor cell killing 32 Shayakhmetov DM, Papayannopoulou T, Stamatoyannopoulos
of a replicating adenoviral vector. Hum Gene Ther 2000; 11: G, Lieber A. Efficient gene transfer into human CD34(+) cells by
379–388. a retargeted adenovirus vector. J Virol 2000; 74: 2567–2583.
14 Kim J, Cho JY, Kim JH, Jung KC, Yun CO. Evaluation of E1B 33 Havenga MJ, Lemckert AA, Grimbergen JM, Vogels R, Huisman
gene-attenuated replicating adenoviruses for cancer gene ther- LG, Valerio D et al. Improved adenovirus vectors for infection of
apy. Cancer Gene Ther 2002; 9: 725–736. cardiovascular tissues. J Virol 2001; 75: 3335–3342.
15 Liu TC, Hallden G, Wang Y, Brooks G, Francis J, Lemoine N et al. 34 Havenga MJ, Lemckert AA, Ophorst OJ, van Meijer M,
An E1B-19kDa gene deletion mutant adenovirus demonstrates Germeraad WT, Grimbergen J et al. Exploiting the natural
tumor necrosis factor-enhanced cancer selectivity and enhanced diversity in adenovirus tropism for therapy and prevention of
oncolytic potency. Mol Ther 2004; 9: 786–803. disease. J Virol 2002; 76: 4612–4620.
16 Degenhardt K, Perez D, White E. Pathways used by adenovirus 35 Segerman A, Atkinson JP, Marttila M, Dennerquist V, Wadell G,
E1B 19K to inhibit apoptosis. Symp Soc Exp Biol 2000; 52: 241–251. Arnberg N. Adenovirus type 11 uses CD46 as a cellular receptor.
17 Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. J Virol 2003; 77: 9183–9191.
Cancer Res 2000; 60: 3689–3695. 36 Gaggar A, Shayakhmetov DM, Lieber A. CD46 is a cellular
18 Piepkorn M. Melanoma genetics: an update with focus on the receptor for group B adenoviruses. Nat Med 2003; 9: 1408–1412.
CDKN2A(p16)/ARF tumor suppressors. J Am Acad Dermatol 37 Sirena D, Lilienfeld B, Eisenhut M, Kalin S, Boucke K, Beerli RR
2000; 42: 705–722. et al. The human membrane cofactor CD46 is a receptor for
19 Fueyo J, Gomez-Manzano C, Alemany R, Lee PS, McDonnell TJ, species B adenovirus Serotype 3. J Virol 2004; 78: 4454–4462.
Mitlianga P et al. A mutant oncolytic adenovirus targeting the Rb 38 Pilder S, Logan J, Shenk T. Deletion of the gene encoding the
pathway produces anti-glioma effect in vivo. Oncogene 2000; 19: adenovirus 5 early region 1b 21,000-molecular-weight polypep-
2–12. tide leads to degradation of viral and host cell DNA. J Virol 1984;
20 Heise C, Hermiston T, Johnson L, Brooks G, Sampson-Johannes 52: 664–671.
A, Williams A et al. An adenovirus E1A mutant that demon- 39 Fleischli C, Verhaagh S, Havenga M, Sirena D, Schaffner W,
strates potent and selective systemic anti-tumoral efficacy. Nat Cattaneo R et al. The distal short consensus repeats 1 and 2 of the
Med 2000; 6: 1134–1139. membrane cofactor protein CD46 and their distance from the cell
Gene TherapyOncolytic adenoviruses for melanoma treatment
M Schmitz et al
13
membrane determine productive entry of species B adenovirus expression of the adenoviral death protein. Hum Gene Ther 2002;
serotype 35. J Virol 2005; 79: 10013–10022. 13: 1859–1871.
40 Stephens C, Harlow E. Differential splicing yields novel 52 Banerjee NS, Rivera AA, Wang M, Chow LT, Broker TR, Curiel
adenovirus 5 E1A mRNAs that encode 30 kd and 35 kd proteins. DT et al. Analyses of melanoma-targeted oncolytic adenoviruses
EMBO J 1987; 6: 2027–2035. with tyrosinase enhancer/promoter-driven E1A, E4, or both in
41 White E, Cipriani R, Sabbatini P, Denton A. Adenovirus E1B submerged cells and organotypic cultures. Mol Cancer Ther 2004;
19-kilodalton protein overcomes the cytotoxicity of E1A pro- 3: 437–449.
teins. J Virol 1991; 65: 2968–2978. 53 Philipson L, Pettersson RF. The coxsackie-adenovirus receptor—
42 Subramanian T, Tarodi B, Chinnadurai G. p53-independent a new receptor in the immunoglobulin family involved in cell
apoptotic and necrotic cell deaths induced by adenovirus adhesion. Curr Top Microbiol Immunol 2004; 273: 87–111.
infection: suppression by E1B 19K and Bcl-2 proteins. Cell 54 Kanerva A, Wang M, Bauerschmitz GJ, Lam JT, Desmond RA,
Growth Differ 1995; 6: 131–137. Bhoola SM et al. Gene transfer to ovarian cancer versus
43 Hemmi S, Geertsen R, Mezzacasa A, Peter I, Dummer R. The normal tissues with fiber-modified adenoviruses. Mol Ther
presence of human coxsackievirus and adenovirus receptor is 2002; 5: 695–704.
associated with efficient adenovirus-mediated transgene expres- 55 Okada N, Masunaga Y, Okada Y, Mizuguchi H, Iiyama S, Mori N
sion in human melanoma cell cultures. Hum Gene Ther 1998; 9: et al. Dendritic cells transduced with gp100 gene by RGD fiber-
2363–2373. mutant adenovirus vectors are highly efficacious in generating
44 Short JJ, Pereboev AV, Kawakami Y, Vasu C, Holterman MJ, anti-B16BL6 melanoma immunity in mice. Gene Therapy 2003; 10:
Curiel DT. Adenovirus serotype 3 utilizes CD80 (B7.1) and 1891–1902.
CD86 (B7.2) as cellular attachment receptors. Virology 2004; 322: 56 Yotnda P, Zompeta C, Heslop HE, Andreeff M, Brenner MK,
349–359. Marini F. Comparison of the efficiency of transduction of
45 Zhang J, Ramesh N, Chen Y, Li Y, Dilley J, Working P et al. leukemic cells by fiber-modified adenoviruses. Hum Gene Ther
Identification of human uroplakin II promoter and its use in the 2004; 15: 1229–1242.
construction of CG8840, a urothelium-specific adenovirus 57 Nemerow GR, Stewart PL. Role of alpha(v) integrins in
variant that eliminates established bladder tumors in combina- adenovirus cell entry and gene delivery. Microbiol Mol Biol Rev
tion with docetaxel. Cancer Res 2002; 62: 3743–3750. 1999; 63: 725–734.
46 Subramanian T, Kuppuswamy M, Gysbers J, Mak S, 58 Suomalainen M, Nakano MY, Boucke K, Keller S, Greber UF.
Chinnadurai G. 19-kDa tumor antigen coded by early Adenovirus-activated PKA and p38/MAPK pathways boost
region E1b of adenovirus 2 is required for efficient synthesis microtubule-mediated nuclear targeting of virus. EMBO J 2001;
and for protection of viral DNA. J Biol Chem 1984; 259: 20: 1310–1319.
11777–11783. 59 Greber UF. Signalling in viral entry. Cell Mol Life Sci 2002; 59:
47 Subramanian T, Kuppuswamy M, Mak S, Chinnadurai G. 608–626.
Adenovirus cyt+ locus, which controls cell transformation and 60 Maizel Jr JV, White DO, Scharff MD. The polypeptides of
tumorigenicity, is an allele of lp+ locus, which codes for a adenovirus. I. Evidence for multiple protein components in the
19-kilodalton tumor antigen. J Virol 1984; 52: 336–343. virion and a comparison of types 2, 7A, and 12. Virology 1968; 36:
48 Hu MC, Hsu MT. Adenovirus E1B 19K protein is required 115–125.
for efficient DNA replication in U937 cells. Virology 1997; 227: 61 Sarnow P, Sullivan CA, Levine AJ. A monoclonal antibody
295–304. detecting the adenovirus type 5-E1b-58Kd tumor antigen:
49 Chiou SK, White E. Inhibition of ICE-like proteases inhibits characterization of the E1b-58Kd tumor antigen in adenovirus-
apoptosis and increases virus production during adenovirus infected and -transformed cells. Virology 1982; 120: 510–517.
infection. Virology 1998; 244: 108–118. 62 Harlow E, Franza Jr BR, Schley C. Monoclonal antibodies
50 Tollefson AE, Ryerse JS, Scaria A, Hermiston TW, Wold WS. The specific for adenovirus early region 1A proteins: extensive
E3-11.6-kDa adenovirus death protein (ADP) is required for heterogeneity in early region 1A products. J Virol 1985; 55:
efficient cell death: characterization of cells infected with adp 533–546.
mutants. Virology 1996; 220: 152–162. 63 Ebbinghaus C, Al-Jaibaji A, Operschall E, Schoffel A, Peter I,
51 Sauthoff H, Pipiya T, Heitner S, Chen S, Norman RG, Rom WN Greber UF et al. Functional and selective targeting of adenovirus
et al. Late expression of p53 from a replicating adenovirus to high-affinity Fcgamma receptor I-positive cells by using a
improves tumor cell killing and is more tumor cell specific than bispecific hybrid adapter. J Virol 2001; 75: 480–489.
Supplementary Information accompanies the paper on the Gene Therapy website (http://www.nature.com/gt).
Gene TherapyYou can also read