BSF Binds Specifically to the bicoid mRNA 39 Untranslated Region and Contributes to Stabilization of bicoid mRNA
←
→
Page content transcription
If your browser does not render page correctly, please read the page content below
MOLECULAR AND CELLULAR BIOLOGY, May 2001, p. 3462–3471 Vol. 21, No. 10
0270-7306/01/$04.00⫹0 DOI: 10.1128/MCB.21.10.3462–3471.2001
Copyright © 2001, American Society for Microbiology. All Rights Reserved.
BSF Binds Specifically to the bicoid mRNA 3⬘ Untranslated
Region and Contributes to Stabilization of bicoid mRNA
RICARDO MANCEBO,1† XIULAN ZHOU,1‡ WENDY SHILLINGLAW,2 WILLIAM HENZEL,2
1,3
AND PAUL M. MACDONALD *
Department of Biological Sciences, Stanford University, Stanford, California 943051; Protein Chemistry Department,
Genentech, Inc., South San Francisco, California 940802; and Section of Molecular Cell and Developmental Biology,
Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin, Texas 787123
Received 4 January 2001/Returned for modification 12 February 2001/Accepted 27 February 2001
The early stages of Drosophila melanogaster development rely extensively on posttranscriptional forms of gene
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
regulation. Deployment of the anterior body patterning morphogen, the Bicoid protein, requires both local-
ization and translational regulation of the maternal bicoid mRNA. Here we provide evidence that the bicoid
mRNA is also selectively stabilized during oogenesis. We identify and isolate a protein, BSF, that binds
specifically to IV/V RNA, a minimal form of the bicoid mRNA 3ⴕ untranslated region that supports a normal
program of mRNA localization during oogenesis. Mutations that disrupt the BSF binding site in IV/V RNA or
substantially reduce the level of BSF protein lead to reduction in IV/V RNA levels, indicating a role for BSF
in RNA stabilization. The BSF protein is novel and lacks all of the characterized RNA binding motifs. However,
BSF does include multiple copies of the PPR motif, whose function is unknown but appears in other proteins
with roles in RNA metabolism.
Many forms of gene regulation occur posttranscriptionally. protein occurs only after fertilization, suggesting that the bcd
These include nuclear functions, such as alternative splicing of mRNA is not translated during oogenesis (10). Activation of
mRNAs, and a variety of processes in the cytoplasm, such as translation is likely to be mediated by cytoplasmic polyadenyl-
regulated stability, localization, translation, and modification ation of the bcd mRNA, which occurs early in embryogenesis
of mRNAs. Most of the cytoplasmic forms of regulation act (32). The bcd mRNA is stable early in embryogenesis but
primarily in controlling the level or distribution of proteins rapidly disappears after about 3 h of growth, revealing a role
encoded by the affected mRNAs and are especially useful in for mRNA stability or instability in its control (38). Each of
two situations. One is when the level of a protein must be these regulated aspects of bcd mRNA activity—mRNA local-
changed very rapidly. Although activation or repression of ization, translational activation, and stability—is mediated by
transcription can modulate the level of gene expression, mech- cis-acting elements present in the bcd mRNA 3⬘ untranslated
anisms that act on mRNAs are more direct and thus can region (UTR) (30, 32, 38). Each element is expected to func-
generate more rapid changes. Cytoplasmic forms of posttran- tion by binding one or more regulatory proteins.
scriptional control are also common for maternal mRNAs. Most of the known regulated mRNAs with prominent roles
Transcripts made by the mother and contributed to the egg in early Drosophila development have been identified through
may be translated long after their synthesis, and the timing of genetic approaches. Similar approaches have proven to be
translational activation or repression can be crucial for normal much less useful for finding the expected regulatory proteins
development (40). Similarly, the localization of some maternal that bind specifically to these mRNAs. Although several genes
mRNAs to specific regions within the egg can be an essential
have been implicated in bcd mRNA localization (36, 37), of
form of regulated gene expression (5).
these only staufen has been shown to encode an RNA binding
Numerous examples of posttranscriptional control of mater-
protein that interacts specifically with bcd mRNA (12). Staufen
nal mRNAs have emerged from the analysis of Drosophila
acts only very late in the localization process (27, 37), so other
melanogaster early development (22). One of these is the ma-
RNA binding proteins must be required. Notably, most of the
ternally contributed bicoid (bcd) mRNA, which encodes a pro-
proteins that bind to cis-acting elements responsible for post-
tein that is largely responsible for organizing the anterior body
transcriptional control of Drosophila maternal mRNAs have
pattern of the embryo (8, 9). Correct deployment of the Bcd
been identified by biochemical rather than genetic strategies
protein in an anterior gradient in the embryo relies on prelo-
calization of bcd mRNA to the anterior pole of the oocyte, (15, 17, 21, 29, 34). One possible explanation for the limited
where it persists into embryogenesis. Accumulation of Bcd success of genetic approaches invokes redundancy. For exam-
ple, multiple proteins could act in recognition of the bcd
mRNA localization signals, and loss of a single binding protein
* Corresponding author. Mailing address: Institute for Cellular and might incur only a subtle defect. In this situation, a mutant
Molecular Biology, Section of Molecular Cell and Developmental Bi- defective for a binding protein would have a weak or imper-
ology, University of Texas at Austin, 2500 Speedway, Austin, TX ceptible phenotype and would not be recovered from simple
78712-1095. Phone: (512) 232-6292. Fax: (512) 232-6295. E-mail: pmac
mutant screens. There is already strong evidence for such re-
@icmb.utexas.edu.
† Present address: Gorilla Genomics, Alameda, CA 94501. dundancy in bcd mRNA localization (27, 28); other regulatory
‡ Present address: Rigel Inc., South San Francisco, CA 94080. processes might also involve redundancy, explaining why ge-
3462VOL. 21, 2001 BSF STABILIZES bcd mRNA 3463
netic approaches have failed to identify many of the regulatory ml, 2 g of leupeptin per ml). Egg chambers were disrupted by Dounce homog-
factors. enization in EB 100 times on ice. The extract was filtered through Miracloth and
spun at ⬃20,000 ⫻ g for 15 min at 0°C in a microcentrifuge. The clear superna-
In previous work we initiated a biochemical approach to tant was removed and recentrifuged. The clear supernatant from the second spin
identify regulatory proteins that bind to the bcd mRNA. We was removed, mixed with cold 50% glycerol to a final concentration of 10%
described a protein, Exl, that binds to a specific region of the glycerol, and frozen in a dry ice-ethanol bath. This preparation yielded approx-
bcd mRNA 3⬘ UTR, and we presented evidence supporting a imately 29.8 mg of protein in 1.78 ml of extract.
(iii) Preparation of small-scale ovary extract. Ovaries were individually dis-
role in mRNA localization (29). Here we have extended that
sected in water at room temperature and placed in cold 1⫻ phosphate-buffered
biochemical approach to find additional binding proteins. We saline (PBS). Isolated ovaries were then washed four times with cold EB-O (50
report the isolation of a protein, BSF, that binds specifically to mM Tris-HCl [pH 7.5 to 7.7], 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100,
the bcd mRNA 3⬘ UTR. Characterization of the function of 0.5 mM DTT, 0.5 mM PMSF, 1 mM benzamidine, 1 g of pepstatin per ml, 1 g
BSF by using both biochemical and genetic assays very strongly of leupeptin per ml). Washed ovaries were homogenized in approximately 2.5
volumes of EB-O on ice in a microcentrifuge tube with a plastic pestle. Extract
suggests that it acts in stabilization of the bcd mRNA during was spun at ⬃20,000 ⫻ g for 15 min at 0 to 4°C in a microcentrifuge. The clear
oogenesis and that it plays a redundant role in this process. supernatant was removed, recentrifuged if any particular matter remained,
Our results add another form of posttranscriptional control to mixed with approximately 0.7 volume of 50% cold glycerol, and frozen in a dry
those known to act through the bcd 3⬘ UTR. Notably, the ice-ethanol bath.
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
sequences required for stabilization during oogenesis are dis- RNA binding assays. UV cross-linking assays were performed as described
(17), except that the binding buffer included 10 mM EDTA, 0.9 mM benzami-
tinct from those involved in destabilization during embryogen- dine, 0.9 g of pepstatin per ml, and 0.9 g of leupeptin per ml. For competition
esis (38), and so the processes appear to be distinct. binding assays (see Fig. 1), increasing amounts of IV/V RNA (trace labeled to
facilitate quantitation) were added to binding reactions at room temperature 5 to
10 min prior to addition of labeled IV/V RNA probe. Proteins for the binding
MATERIALS AND METHODS
assays were from ovarian extracts (described above) or synthesized from cDNAs
DNAs. DNA fragments corresponding to wild-type IV/V, IV, and V RNAs in coupled transcription and translation extracts (Promega).
(27), all subdomains of the bcd mRNA 3⬘ UTR (see Fig. 1), were subcloned into Purification of BSF. Ovary extract from the large-scale preparation described
transcription vectors for preparation of RNA probes (p2865, p5008, and p2866). above was fractionated on a 1-ml Q-Sepharose Hi-Trap column (Pharmacia)
Linker scanning (LS) mutants were constructed by standard PCR methods using using an Econo Low-Pressure Chromatography System (Bio-Rad) at a flow rate
oligonucleotide primers designed to replace adjacent 10-nucleotide (nt) seg- of 1 ml/min. The column was loaded and washed with buffer A (20 mM Tris-HCl
ments of IV/V with the sequence GAAUCGAUUC. LS2 replaces nt 4399 to [pH 8.0], 10% glycerol, 0.5 mM DTT, 0.5 mM PMSF) also containing 100 mM
4408 (coordinates from GenBank accession number X51741) and LS3 replaces NaCl. Proteins were eluted in buffer A containing 298 mM NaCl. Eluted frac-
nt 4409 to 4418, and the series continues with 10-nt substitutions up to LS27, tions were tested for P150 binding activity using the UV cross-linking assay
which replaces nt 4649 to 4658. LS15 replaces UAUUUUCAAU (nt 4529 to described above.
4538). All mutants were constructed in transcription vectors to allow synthesis of Fractions demonstrating BSF activity from the Q-Sepharose column were
sense RNAs for use as probes in RNA binding assays. Individual LS mutants, incubated with 65 l of Reactive 4 Blue Dye resin at 4°C for approximately 4.5 h.
flanked by XbaI restriction sites, were subcloned into the P transformation vector The supernatant was removed and the resin was washed with 100 mM NaCl, 20
p2405. This vector includes the osk promoter, the green fluorescent protein mM Tris-HCl (pH 8.0), 10% glycerol, 0.5 mM DTT, 0.5 mM PMSF, 1 mM
(GFP) coding region, and a short 3⬘ UTR with a polyadenylation signal; the benzamidine, 1 g of pepstatin per ml, and 1 g of leupeptin per ml. Proteins
cloning site is within the 3⬘ UTR (26). The wild-type version is P[gfpIV/V], and bound to the dye resin were eluted with IV/V RNA at 4°C on a rotator for
the mutants are, for example, P[gfpIV/VLS15]. For experiments to test the approximately 1.5 h. The elution solution contained 10 g of IV/V RNA (made
function of BSF, a second reporter transgene bearing the IV/V localization signal using the T7-MEGAshortscript transcription system from Ambion) and 350 g
was also used. It makes use of the bicoid promoter and includes a lacZ sequence of yeast tRNA in 260 l of elution buffer (approximately 2.2 mM MgCl2, 6.5 mM
tag (27). Tris-HCl [pH 7.5], 11 mM EDTA, 0.5 mM PMSF, 1 mM benzamidine, 1 g of
The cDNA clone SD10676 of the Berkeley Drosophila Genome Project pepstatin per ml, 1 g of leupeptin per ml). Eluted proteins were separated by
(BDGP) was obtained from Research Genetics (Birmingham, Ala.). Restriction sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
fragments of the cDNA were subcloned for DNA sequencing. A deletion deriv- electroblotted to a polyvinylidene difluoride membrane (ProBlott; Applied Bio-
ative of bsf encoding the N-terminal 1,007 amino acids was subcloned into the systems).
pET3a vector for expression in Escherichia coli. Protein sequencing. The 150-kDa protein band was excised from the polyvi-
Antibodies. The N-terminal 1,007 amino acids of BSF were expressed in E. nylidene difluoride membrane and wetted with 1 l of methanol. The band was
coli, partially purified, and used as an immunogen for production of antiserum in reduced and alkylated with isopropylacetamide (18) followed by digestion in 20
rats. Antiserum to the BSF protein was tested by Western blot analysis of TNT l of 0.05 M ammonium bicarbonate containing 0.5% Zwitergent 3-16 (Calbio-
reactions (Promega) expressing either BSF or a control protein in rabbit reticu- chem) with 0.2 g of trypsin (Frozen Promega Modified) at 37°C for 17 h (23).
locyte lysates. A band of approximately 150 kDa was detected in ovary extracts Peptides generated by trypsin were subjected to collision-induced dissociation
and in the lysates expressing BSF and was absent in the control lysate (R. in an ion trap mass spectrometer (LCQ; Finnigan MAT). A 1-l aliquot (5%) of
Mancebo and P. M. Macdonald, unpublished). the tryptic digest was loaded onto a 75-m inside diameter, 360-m outside
Drosophila ovarian extracts. (i) Large-scale isolation of ovaries. Embryos from diameter, 20-cm length of fused silica capillary packed with 15 cm of POROS
W1118 flies were collected overnight onto egg-laying plates in large fly houses and 10R2 reverse-phase beads (PerSeptive Biosystems). Peptides were eluted with 15
were seeded into larval containers. The larvae and adults that emerged were fed min of acetonitrile gradient at a flow rate of 200 nl/min as previously described
yeast paste (autoclaved to reduce the amount of proteolysis that might occur (3).
during ovary extract preparation if active yeast were present in the gut). The Peptide masses and selected b and y series fragment ions were used to search
adults were homogenized in approximately 10 liters of IB (20 mM Tris-HCl [pH an in-house (Genentech) protein and DNA sequence database with an enhanced
7.5], 100 mM NaCl, 1 mM EDTA) with a blender using short pulses at low speed. version of the FRAGFIT program (2, 16).
The total amount of flies homogenized was 215.5 g. The homogenate was first bsf genetics. The P element stock l(2)k07109, generated and characterized by
filtered through a 500-m mesh membrane and collected onto a 70-m mesh the BDGP, was obtained from Amy Beaton at the BDGP. The stock carries two
membrane. The filtrate that was retained on the 70-m mesh was then filtered P element insertions, one in 25F1-2 and one in 36E3-4. We used complementa-
through a 150-m mesh and collected onto a 60-m mesh membrane, followed tion tests with Df(2L)M36F-S5 to show that the 36E P insertion is not lethal.
by washes with IB. The settled volume of filtrate was 1.5 ml and was enriched for Because the 36E P insertion reduces but does not eliminate BSF protein (see Fig.
all stages of egg chambers. 6A), we initiated efforts to use this insertion to make stronger alleles of bsf.
(ii) Preparation of large-scale ovary extract for purification. The filtrate from However, recombination and complementation tests strongly suggest that the
the large-scale isolation of ovaries was washed four times with 20 ml of EB (20 36E P element of l(2)k07109 is no longer w⫹, rendering it useless for any screen
mM Tris-HCl [pH 8.0], 100 mM NaCl, 2 mM dithiothreitol [DTT], 1 mM for stronger alleles of bsf that relies on loss of w⫹. Consequently we halted our
phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine, 2 g of pepstatin per efforts to obtain stronger alleles.3464 MANCEBO ET AL. MOL. CELL. BIOL.
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
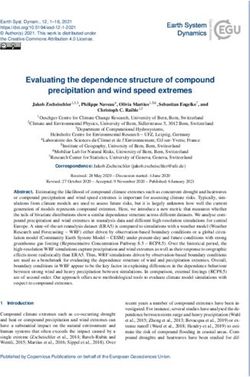
FIG. 1. Proteins that bind to IV/V RNA. A schematic diagram of the predicted secondary structure of the bcd mRNA 3⬘ UTR is shown at the
left, with individual domains indicated by roman numerals. The subdomains of the RNA used in this work, IV/V, IV (created by site-directed
mutagenesis to remove V; see reference 27), and V, are shown below. UV cross-linking was used to monitor binding of Drosophila ovarian proteins
to 32P-labeled RNA probes (identified above each lane). Individual binding proteins are identified by approximate molecular size at the left
(migration of size standards noted at the right). For the competition binding experiments, 0.1-, 1-, 12-, or 122-fold molar excess cold IV/V RNA
was added to the binding reactions. Subdomains of IV/V RNA (IV and V) or a mutant IV/V that is defective in RNA localization in vivo (G4496U)
(27) were used to better define binding specificities. Two proteins, p55 and p70, bind sites in V based on their strong binding to IV/V and V and
weak or nondetectable binding to IV. Three proteins, p58, p105, and p160, bind to all probes tested. Two proteins, p80 and BSF, require the intact
IV/V RNA for strong binding. None of the binding interactions is noticeably affected by the G4496U point mutation. SL, stem-loop.
Determination of mRNA levels in transgenic flies. Females bearing individual were mounted in Vectashield (Vector Labs) and examined by confocal micros-
LS mutant transgenes were fattened, their ovaries were dissected, and their RNA copy using a Leica TCS SPII microscope.
was isolated (25). For each reaction, 10 g of nucleic acid was mixed with labeled Specificity of the antibodies was determined by comparing fluorescence levels
RNA probes from both the gfp and bcd genes to compare the levels of LS mutant in wild-type and bsf1/Df(2L)M36F-S5 transheterozygous ovaries. To allow a di-
IV/V RNAs with endogenous bcd mRNA, respectively, in multiprobe RNase rect comparison of the two genotypes, the wild-type flies also carried a transgene
protection assays (41). expressing GFP fused to asparaginyl tRNA synthetase, which is dispersed
To test the role of BSF in stabilization of bcd mRNA, Df(2L)M36-S5/CyO throughout the cytoplasm (R. Mancebo and P. M. Macdonald, unpublished).
Dp(2;2)M(2)m⫹ females were crossed to l(2)k07109/CyO; P[gfpIV/V]/TM2 The wild-type and bsf mutant flies were mixed and processed (dissected, fixed,
males. Progeny females of the genotype Df(2L)M36-S5/l(2)k07109; P[gfpIV/V]/⫹ and stained) together, with the genotypes distinguished by the presence or
were fattened in yeasted vials for 2 to 4 days. Ovaries were dissected, RNA was absence of GFP fluorescence.
isolated, and the levels of reporter (gfpIV/V) and endogenous bcd mRNAs were Nucleotide sequence accession number. Our sequence of the SD10676 cDNA
determined by RNase protection assay (RPAIII; Ambion). As controls, RNAs has been deposited at GenBank with accession number AF327844.
from the sibling genotypes of Df(2L)M36-S5/CyO; P[gfpIV/V]/⫹, l(2)k07109/
CyO; P[gfpIV/V]/⫹, and Df(2L)M36-S5/CyO; ⫹/TM2 were also tested. RNase
protection assays were quantitated by phosphorimaging (Molecular Dynamics) RESULTS
using three independent sets of assays. These experiments were repeated using To search for proteins that may regulate the activity or
the bcd⫹lacZIV/V reporter (27) with similar results.
distribution of bcd mRNA we focused on the IV/V region (Fig.
Immunolocalization of BSF. Adult females were fattened and ovaries were
dissected in PBS. Ovaries were disrupted by being rapidly pipetted through a 1), a 271-nt portion of the bcd 3⬘ UTR that supports a normal
drawn-out pasteur pipette and were fixed for 30 min in a 1.5-ml microcentrifuge pattern of mRNA localization during oogenesis (27). Unlike
tube containing 200 l of PBS, 40 l of 37% formaldehyde, and 1 ml of heptane. the complete 3⬘ UTR, the IV/V RNA lacks redundant infor-
The samples were washed in several changes of PBT (PBS plus 0.1% Triton mation for the initial step of bcd mRNA localization. Specifi-
X-100) and 5% goat serum, incubated overnight in rat anti-p150 diluted 1:1,000
in PBT and 1% goat serum, washed five to seven times in PBT and 5% goat
cally, the localization activity of IV/V can be greatly reduced by
serum, incubated for 2 h in Cy5-labeled goat anti-rat secondary antibodies a point mutation (G4496U), while the same mutation has only
(Jackson Immunochemicals) diluted 1:600 in PBT, and washed in PBT. Samples a subtle and transient effect on the activity of the complete 3⬘VOL. 21, 2001 BSF STABILIZES bcd mRNA 3465
UTR. The absence of functional redundancy is a prerequisite
for experiments in which an attempt is made to correlate a
protein binding site with a biological activity.
Extracts prepared from Drosophila ovaries were tested for
the presence of proteins that bind to IV/V RNA using a UV
cross-linking assay. A number of proteins bind under these
conditions, and all those larger than 50 kDa are identified in
Fig. 1. The assays were also performed in the presence of
increasing amounts of competitor RNA as an initial test for
binding specificity. Most of the bands detected in the assay are
unaffected by addition of the competitor, but the binding of
four proteins, p55, p70, p80, and BSF, is clearly reduced. To
explore a possible role for any of the proteins in bcd mRNA
localization, RNA probes corresponding to the isolated parts
of IV/V (predicted stem-loops IV and V) (Fig. 1) were used in
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
separate binding assays; RNAs IV and V have no localization
activity in vivo and thus may fail to bind one or more localiza-
tion factors (27). Many proteins bind equally well to all probes.
Two proteins, p55 and p70, bind to V RNA but not IV RNA,
suggesting that they recognize sites contained entirely within
V. Finally, p80 and BSF bind much better to IV/V RNA than
to either of the isolated parts (which do not support mRNA FIG. 2. BSF binding to LS mutant IV/V RNAs. LS mutant RNA
localization) and are thus the best candidates to act in mRNA probes were used for UV cross-linking assays. Each panel shows a
localization. To explore further a possible role for the cross- separate experiment testing the binding of BSF to a subset of the
linking proteins in bcd mRNA localization, a binding assay was mutants. LS15 displays the most severe reduction in BSF binding (the
done with a point-mutated IV/V RNA (G4496U) that inter- autoradiographic exposure is longer in this panel than in others; note
the intensity of the wild-type [wt] band). Several mutants, including
feres with mRNA localization in vivo (27). The G4496U mu- LS19 and LS20, also show reduced BSF binding, but to a lesser extent.
tation had no effect on the binding of any of the proteins Some of the mutants display enhanced binding to BSF. Although we
detected in this assay. Although this result does not rule out do not know the reason for this effect, it could reflect subtle differences
involvement of any of the binding proteins in bcd mRNA in RNA folding, and we have observed similar effects for other RNA
binding proteins (17).
localization, it does suggest that other roles may be more likely.
Here we characterize one of the proteins, BSF, and we de-
scribe experiments that assess its function. Our data argue very
strongly for a role in stabilization of bcd mRNA, so we refer to phenotypic description of the full set of linker scan mutants
this protein as bicoid mRNA stability factor or BSF. will be published elsewhere). The simplest interpretation of
Identification of a stability element within the bcd 3ⴕ UTR our results is that the LS15 mutant destabilizes the IV/V RNA.
that is recognized by BSF. Our strategy for testing the role of Although we cannot exclude alternate explanations that invoke
BSF was to first identify mutations in IV/V RNA that interfere low-probability events (such as alteration of the transgene dur-
with BSF binding in vitro and then to determine the conse- ing transposition into the Drosophila genome or selective and
quences of these same mutations in vivo. We used LS mu- consistent targeting of the transgene to regions of the genome
tagenesis to create a series of 27 mutants that collectively alter that interfere with its expression), data presented below are
most of the 271 nt of IV/V. Each mutant replaces a 10-nt fully consistent with the conclusion that the LS15 mRNA is
segment of IV/V with a synthetic sequence (see Materials and unstable and that instability is the consequence of the defect in
Methods). Wild-type and mutant IV/V RNAs were used as BSF binding.
probes in binding assays, as shown in the three panels of Fig. 2. Biochemical purification of BSF. To initiate a biochemical
Each panel represents a separate experiment, and band inten- and genetic analysis of BSF, we purified the protein from
sities should be compared to the wild-type control from the isolated Drosophila ovaries. The purification is outlined in Fig.
same experiment. One mutant RNA, LS15, is most severely 4A and is described in detail in Materials and Methods. For the
impaired in BSF binding. Several others (e.g., LS19 and LS20) penultimate step of purification, proteins were bound to Re-
are also impaired but to a lesser extent. Almost all of the same active 4 Blue Dye and specifically eluted using IV/V RNA.
LS mutants were also tested in vivo. Each mutant was intro- Chromatography on the dye column was first performed on an
duced into a reporter construct (27), transgenic fly strains were analytical scale, by eluting with radiolabeled RNA and testing
established, and patterns of mRNA localization in transgenic fractions for the presence of BSF by a UV cross-linking assay
ovaries were monitored by in situ hybridization. For the LS15 (Fig. 4B, lanes 4 through 8). The separation was repeated at a
mutant, no localized reporter mRNA was detected in any of preparative scale using unlabeled IV/V RNA. Silver staining of
four independent transgenic fly stocks, and the underlying eluted fractions separated by SDS-PAGE revealed a single
cause was unique among all mutants tested: the LS15 mutant prominent protein band in the expected size range as well as a
fails to accumulate any mRNA (Fig. 3). In comparison, the number of other smaller proteins (Fig. 4B, lane 9). The can-
LS11 mutant, which is completely defective in mRNA local- didate BSF protein was excised from the gel and processed for
ization, retains normal levels of transgene mRNA (a complete sequencing.3466 MANCEBO ET AL. MOL. CELL. BIOL.
the amino terminus and three copies dispersed over the car-
boxyl-terminal half of the protein. The PPR motif, which is
usually about 35 amino acids long, has no assigned function but
has already been identified in over 200 proteins that are widely
represented in plant organelles (33). Two proteins that contain
the motif have been characterized genetically, and each plays a
role in RNA metabolism (4, 14, 31). Notably, the PPR-con-
taining PET309 protein has been shown to act in either pro-
cessing or stabilization of certain RNAs in yeast (31). Based on
these observations, BSF is a member of a new protein family
that may have a common function in RNA metabolism.
Sequence comparisons of BSF with those of the GenBank
database identify two proteins that are most closely related, a
human leucine-rich protein of unknown function (expectation
[E] value of ⬍10⫺136) and a predicted Drosophila protein (E ⬍
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
10⫺68). Both proteins also contain multiple copies of the PPR
motif, although the extensive homology between BSF and the
leucine-rich protein is not limited to these repeated structural
elements.
Despite the biochemical evidence that BSF is an RNA bind-
ing protein, none of the known RNA binding motifs are pres-
ent in the protein. Thus, BSF appears to define a novel class of
RNA binding protein.
Mutations that reduce the level of BSF in the cell lead to a
FIG. 3. Flies carrying LS15 transgenes fail to accumulate IV/V reduction of IV/V RNA. The cytological map position of the bsf
RNA. The autoradiograph shows RNase protection assays that probe gene is on the left arm of the second chromosome at 36E3-4
the levels of both endogenous bcd RNA and transgenic gfp reporter (BDGP). A search of the BDGP database revealed that none
RNA in the same reaction (designated at the left). The two gfp bands of the P element insertion mutants (35) whose exact chromo-
presumably reflect RNase cleavage within the probe, a common oc-
somal position has been determined by DNA sequencing lies in
currence. RNA samples were from females carrying a wild-type gfp
IV/V reporter (IV/V wt) or LS mutant gfp IV/V reporter or from 0- to or near the bsf gene. However, there are other P element
24-h embryos with no transgene (lane labeled with a minus). The latter insertions in the same region that have not been mapped so
lane reveals background bands (indicated with arrowheads) that mi- precisely. These mutants were obtained, and heterozygous flies
grate near the gfp signals but are not from gfp, being present in RNA were tested for BSF protein levels by Western blot analysis.
samples lacking gfp RNA. Neither of two independent transgenic
stocks of the LS15 mutant shows detectable reporter mRNA (there is One mutant stock, l(2)k07109, displays a reduced level of BSF.
no signal at the positions of the gfp reporter bands, shown more clearly The l(2)k07109 chromosome carries two P element insertions
in the lower panel, which is a longer exposure of a part of the auto- on the second chromosome, one in the bsf region and one in
radiograph shown in the upper panel). Other LS mutant transgenic 25F. The latter insertion is responsible for the lethal pheno-
stocks display some variation in transcript levels, as seen here for two
type, as the BDGP has shown that l(2)k07109 is lethal in trans
independent LS20 stocks, but the gfp RNA can always be detected, in
contrast to what is observed for the LS15 stocks. to a deficiency removing 25F, and we found that l(2)k07109 is
viable in trans to Df(2L)M36F-S5, a deficiency that removes all
of 36E. The viability of l(2)k07109/Df(2L)M36F-S5 flies al-
lowed us to test them for BSF protein levels, which are very
Amino acid sequences were obtained from 11 peptides from substantially reduced (Fig. 6A). Thus, the 36E P insertion of
the purified BSF protein. Six of these peptides correspond to l(2)k07109 reduces the level of BSF protein, a common phe-
three expressed sequence tags (ESTs) from the BDGP; we notype for P element mutants, and we refer to the P insertion
subsequently found all three ESTs to be derived from the same as bsf1. Our efforts to use the bsf1 flies to generate stronger
mRNA, which also contains sequences corresponding to three alleles have failed for reasons outlined in Materials and Meth-
of the other sequenced peptides. One of the EST cDNAs ods. Nevertheless, even in the absence of a null allele the
(SD10676) was translated in vitro, and the reaction products bsf1/Df(2L)M36F-S5 transheterozygotes may provide a partial
were used in the UV cross-linking assay. As shown in Fig. 4C, loss-of-function phenotype for bsf.
the cDNA encodes a protein of about 150 kDa that binds IV/V Flies transheterozygous for bsf1 and Df(2L)M36F-S5 are vi-
RNA, supporting the conclusion that the cloned gene is bsf. able and fertile, with no obvious morphological defects in
Genetic analysis (below) confirms this conclusion. oogenesis. When eggs from such females are fertilized by wild-
BSF contains multiple copies of the PPR motif. The cDNA type or bsf1 sperm, they progress normally through embryo-
SD10676, which encodes BSF, was completely sequenced. The genesis and display no cuticular pattern defects (development
single large open reading frame predicts a protein of 1,412 is arrested later for bsf1/bsf1 individuals because of the 25E
amino acids and includes 9 of 11 peptide sequences obtained lethal mutation on the chromosome). As a first test for a
from the purified BSF protein (Fig. 5). The most notable molecular defect in the ovaries of bsf1/Df(2L)M36F-S5 fe-
feature of the BSF sequence is the presence of seven copies of males, we examined the level of endogenous bcd mRNA but
the PPR motif, with four copies adjacent to one another near found no substantial difference relative to the wild type (dataVOL. 21, 2001 BSF STABILIZES bcd mRNA 3467
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
FIG. 4. Purification of BSF. (A) Outline of the purification. SL, stem-loop. (B) Analysis of BSF during the purification steps. All lanes, except
lane 9, show UV cross-linking assays. Lanes 1 through 3 show the purification of BSF away from other RNA binding activities during Q-Sepharose
fractionation. Lanes 4 through 8 show the elution of BSF from Reactive 4 Blue Dye. All of the BSF binding activity is retained on the column (lanes
4 and 5) and can be eluted with labeled IV/V RNA (lanes 6 through 8). Lane 9 shows a silver-stained gel of the eluted proteins from the Reactive
4 Blue Dye. An arrowhead indicates the BSF protein. FT, flowthrough. (C) A cDNA identified from peptide sequences from purified BSF encodes
a 150-kDa protein that binds IV/V RNA. Each lane shows the result of a UV cross-linking assay with IV/V RNA probe and the following extracts:
lane 10, in vitro translation (IVT) of the putative bsf RNA in rabbit reticulocyte lysates; lane 11, ovary extract; lane 12, in vitro translation of
luciferase RNA in rabbit reticulocyte lysates. Proteins synthesized by in vitro translation (lanes 10 and 12) are not purified, and the background
binding activities are those from the translation system.
not shown). However, the apparent mRNA instability pheno- mRNA stabilization role suggested for BSF by the LS mutant
type associated with LS15 (the LS mutant defective in BSF analysis.
binding) is detected for transgenes containing only the IV/V BSF appears in particles within the cytoplasm during oo-
portion of the bcd mRNA 3⬘ UTR (Fig. 3), while deletion from genesis. The subcellular distribution of BSF protein was de-
the complete bcd 3⬘ UTR of the region corresponding to LS15 termined by immunofluorescent detection in whole-mount
has no substantial effect on mRNA levels (mutants ⌬15 and ovaries (Fig. 7). At all stages of oogenesis the protein is cyto-
⌬16 of the gene are described in reference 28). Thus, LS15 plasmic. During the previtellogenic stages of oogenesis BSF is
could block the action of only a single component of a redun- present in both the nurse cells and the oocyte at similar levels
dant stabilizing system, and reduction of BSF activity would (Fig. 7A and B). Within the nurse cells BSF appears primarily
only be detected when redundancy is eliminated. Accordingly, in regions surrounding the nuclei, and within these regions the
we tested the level of the reporter mRNA bearing the wild- protein is often concentrated in a punctate pattern (Fig. 7A).
type IV/V RNA in flies transheterozygous for bsf1 and As oogenesis proceeds (Fig. 7C), the tight association of BSF
Df(2L)M36F-S5. Compared to control flies, the bsf mutant flies with nurse cell nuclei is lost. The particulate appearance of
display a consistent three- to fivefold reduction in the level of BSF is enhanced, but the particles are more evenly dispersed
wild-type IV/V RNA (Fig. 6B). We have no direct evidence throughout the cytoplasm of the nurse cells. In the oocyte the
that proves a mechanism by which a reduction in BSF levels BSF levels are reduced relative to the nurse cells. At no time
reduces the level of IV/V RNA. Nevertheless, the fact that does BSF appear to be concentrated at sites of bcd mRNA
BSF binds to sequences within IV/V RNA argues for a post- accumulation, at either the apical regions of the nurse cells or
transcriptional role, an interpretation consistent with the the anterior margin of the oocyte.3468 MANCEBO ET AL. MOL. CELL. BIOL.
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
FIG. 5. Predicted BSF protein sequence. The sequence shown is that predicted by conceptual translation of the SD10676 EST cDNA. Nine
peptides sequenced from purified BSF are overlined (note that two peptides, LSSGELEPVPLPNSGK and ILNSLAEAGQPER, are each split
between two lines). The seven iterations of the PPR motif are indicated by black boxes with white text. The protein corresponds to CG10302 of
the proteins predicted from analysis of the Drosophila genome sequence (1) (http://www.fruitfly.org/annot/). There are several differences between
our experimental data and the characteristics of the predicted protein. The most significant is that our sequence includes an additional 137 amino
acids at the amino terminus. One of the sequenced peptides lies within this region, confirming that this part of the cDNA is indeed translated.
DISCUSSION suggested by known mechanisms of mRNA stabilization. Spe-
The bcd mRNA has long served as a model example of the cifically, binding of the iron response element protein to se-
importance and variety of posttranscriptional regulatory events quences in the transferrin receptor mRNA 3⬘ UTR blocks
that can be mediated by the 3⬘ UTR. Sequences that direct the endonucleolytic attack and thus stabilizes the mRNA (7). It is
subcellular localization, translation, and embryonic degrada- possible that the LS15 mutant disrupts the BSF binding site but
tion of bcd mRNA have all been found within the 3⬘ UTR. not a nearby nucleolytic cleavage site and thus confers insta-
Here we have identified an additional function, mRNA stabi- bility. In contrast, the larger deletion mutants that do not affect
lization during oogenesis. Our data indicate that stabilization stability of the bcd 3⬘ UTR (⌬15 and ⌬16 of the gene described
of the IV/V domain of the bcd 3⬘ UTR is achieved by interac- in reference 28; 45 and 54 nt deleted, respectively) might elim-
tion of the BSF protein with a cis-acting stability element in the inate both the protection and cleavage elements, making them
RNA. Three lines of evidence support this model of BSF resistant to targeted degradation. Distinguishing among these
action. First, mutation of the stability element eliminates ac- and other possible explanations will require more detailed
cumulation of RNA. Second, BSF binding is greatly reduced by analysis of the cis-acting elements.
the same mutation in the RNA stability element. And third, a Interaction of BSF with IV/V RNA. BSF binds IV/V RNA
mutant with significantly reduced expression of BSF displays a significantly better than either IV or V RNA alone. IV/V may
reduction in the level of IV/V RNA. Taken together, these be a better binding substrate because it contains more itera-
data make a very strong case for BSF-mediated stabilization of tions of a repeated binding site. Alternatively, the binding site
bcd mRNA. may consist of elements from both the IV and V regions, or
Redundancy. The consequence of mutating either the IV/V presentation of the binding site may require a structure that
RNA stability element or bsf is the elimination or reduction, only the complete IV/V RNA can adopt. In this case the
respectively, of the reporter mRNA bearing the IV/V 3⬘ UTR. weaker binding to the isolated parts could represent nonspe-
In contrast, mutation of bsf has no discernable effect on en- cific binding. Although several LS mutants do reduce BSF
dogenous bcd mRNA. Similarly, deletion of the LS15 region binding, as might be expected if each disrupts one copy of a
from the full bcd 3⬘ UTR is tolerated, and the deletion mutant repeated binding site, the fact that the LS15 mutant has a more
RNAs are readily detectable (28). The striking context depen- dramatic effect argues against simple repetition of equivalent
dence of mutating either the cis or trans components of this binding sites. The other option, that BSF recognizes a site
RNA stabilization system suggests that there is redundancy in whose composition or formation requires the intact IV/V, ap-
the stabilization process: sequences outside IV/V are sufficient pears to be more consistent with our data, although the options
for stabilization, and another factor(s) can perform the same are not mutually exclusive. The notion that IV/V is structured
function as BSF. This redundancy is not surprising given the is not new, and there are now several studies either suggesting
redundancy already demonstrated for localization of bcd or demonstrating that at least part of this region must adopt a
mRNA (27). Indeed, the IV/V subdomain of bcd RNA was specific folding for mRNA localization (13, 24, 26).
used in this work because it lacks the mRNA localization The RNA binding domain of BSF appears to be novel, as
redundancy of the full 3⬘ UTR. none of the known RNA binding motifs appear in the protein.
An alternative explanation for the observation that a muta- The single type of recognizable domain, the PPR motif, is also
tion in the RNA stabilization element leads to a reduction in found in other proteins that act in RNA metabolism (14, 31),
the level of IV/V RNA while a deletion of the stabilization and one or more of the seven copies of this motif in BSF could
element from the full bcd 3⬘ UTR appears to have no affect is contribute to RNA binding. However, BSF must also fulfill itsVOL. 21, 2001 BSF STABILIZES bcd mRNA 3469
FIG. 6. A mutant with a reduced level of BSF protein has reduced
levels of IV/V RNA. (A) Ovaries from wild-type and bsf1/Df(2L)
M36F-S5 females were tested for levels of BSF RNA binding activity
(upper panel) and BSF protein (middle panel). RNA binding was
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
monitored by a UV cross-linking assay. The binding to p160 (see Fig.
1) serves as a control to demonstrate that similar amounts of wild-type
and mutant extracts were used for the assays. Levels of BSF protein in
wild-type and mutant extracts were determined by Western blot anal-
ysis using antibodies raised against a bacterially expressed part of BSF.
Loading controls are provided by the light background band at the top
of the BSF blot and reprobing of the blot with anti-tubulin antibodies
(bottom panel). (B) RNase protection assays of ovarian RNA levels for
endogenous bcd mRNA and the reporter gfp mRNA bearing the wild-
type IV/V domain of the bcd 3⬘ UTR. The endogenous bcd mRNA is FIG. 7. Distribution of BSF in ovaries. Anti-BSF antibodies were
not affected by reduction in the level of BSF and is used to normalize used to detect BSF protein in whole-mount ovaries. Signal specificity
the amounts of RNA used for each assay. RNAs are from ovaries of was confirmed by comparison of wild-type ovaries (shown) with bsf1/
flies with the genotypes indicated at the top (a plus sign is used to Df(2L)M36F-S5 ovaries (see Materials and Methods). (A) Egg cham-
indicate bsf1, as carried on a Sp or CyO chromosome). Df and a minus ber showing the concentration of BSF in a punctate pattern surround-
sign indicate the deficiency and P insertion mutants, respectively ing nurse cell nuclei. A similar distribution is observed in the somatic
[Df(2L)M36-S5 and bsf1], and the reporter RNA transgene is indicated layer of follicle cells that surround the nurse cells and oocyte, with
by IV/V. The last lane is a control to show that no gfp RNA signal is differences in the degree of perinuclear concentration among different
detected in the absence of the reporter transgene. Flies transheterozy- follicle cells. (B) Posterior part of an egg chamber in which the level of
gous for Df(2L)M36-S5 and bsf1 (third lane) show a consistent 3- to BSF in the oocyte (oo) is similar to that in the nurse cells (nc). (C)
5-fold reduction in reporter RNA levels (as determined by phosphor- Vitellogenic stage egg chamber. BSF is now present at lower levels in
imaging analysis; see Materials and Methods) relative to the control the oocyte. Within the nurse cells the punctate distribution of BSF
flies (first two lanes). Similar results were obtained using a different persists, but the concentration of BSF at the periphery of nurse cell
transgene bearing the IV/V localization signal but driven by the bcd nuclei is lost. This egg chamber displays a preferential accumulation of
promoter. BSF in the nurse cells closest to the oocyte; this pattern is common but
not universal. The follicle cells continue to show variable concentra-
tions around the nuclei.
function of stabilizing the IV/V RNA, and the PPR motifs
could act in this process. Identification and analysis of the BSF mRNA, experiments typically not done without some prelim-
RNA binding domain may resolve these issues. inary indication that a regulatory sequence exists. A notable
mRNA stability in Drosophila development. Control of exception is the ␣2-globin mRNA, which is extraordinarily
mRNA stability serves an important role in Drosophila devel- stable in erythroid cells. A naturally occurring mutation causes
opment. Multiple transcripts with roles in development of the destabilization, although the mutation does not directly affect
adult peripheral nervous system are normally destabilized the sequences conferring stability. Instead, the stop codon is
posttranscriptionally by conserved 3⬘ UTR regulatory se- inactivated, allowing ribosomes to displace stabilizing factors
quences (19, 20). In the embryo the transcripts of the pair-rule from the 3⬘ UTR of the mRNA (39). The best examples of
genes have extremely short half-lives, a property that contrib- actively stabilized mRNAs in Drosophila are those where sta-
utes to their restriction to the apical cytoplasm at the periphery bilization is a local phenomenon; the retention of an mRNA in
of the blastoderm (11). Additionally, a number of maternal only one part of the embryo clearly suggests that the mRNA is
mRNAs, including bcd, are programmed for rapid turnover either moved or selectively degraded or stabilized. Localized
during the first few hours of embryogenesis, with some varia- stabilization was first demonstrated for the posteriorly local-
tion in the timing of elimination (6). Recent studies on the ized hsp83 mRNA, and it now appears that the same mecha-
timing of degradation and the cis-acting elements acting in this nism may contribute to the localization of other maternal
process for several such mRNAs have revealed the existence of mRNAs at the posterior pole of the embryo (6). The action of
at least two distinct degradation pathways (6). For some ma- differential mRNA stability in mRNA localization raises the
ternal mRNAs instability elements have been identified, al- question of whether mRNA instability may also act in bcd
though no common features have yet emerged (6, 34, 38). mRNA localization. Although we cannot exclude a minor con-
There are fewer examples of mRNAs that are actively sta- tribution of such a mechanism to localization of the bcd
bilized, perhaps in part because evidence for stabilization mRNA, we have found no supporting evidence. Mutations in
would most likely come from directed mutagenesis of the either the complete bcd 3⬘ UTR or the isolated IV/V RNA that3470 MANCEBO ET AL. MOL. CELL. BIOL.
interfere with localization typically have no measurable effect 9. Driever, W., and C. Nüsslein-Volhard. 1988. A gradient of bicoid protein in
Drosophila embryos. Cell 54:83–93.
on stability (Fig. 3) (28). The only possible exception is the 10. Driever, W., V. Siegel, and C. Nüsslein-Volhard. 1990. Autonomous deter-
destabilized LS15 mutant, for which localization cannot be mination of anterior structures in the early Drosophila embryo by the bicoid
monitored. Despite the apparent differences in the roles for morphogen. Development 109:811–820.
11. Edgar, B. A., M. P. Weir, G. Schubiger, and T. Kornberg. 1986. Repression
instability, it is possible that the same machinery may be used. and turnover pattern fushi tarzu RNA in the early Drosophila embryo. Cell
Bashirullah and coworkers (6) have narrowly mapped the 47:747–754.
hsp83 mRNA protection element to a 106-nt segment of the 3⬘ 12. Ferrandon, D., L. Elphick, C. Nüsslein-Volhard, and D. St. Johnston. 1994.
Staufen protein associates with the 3⬘UTR of bicoid mRNA to form particles
UTR, and we have compared this to the segment of bcd that move in a microtubule-dependent manner. Cell 79:1221–1232.
mRNA that, on the basis of the LS15 phenotype, includes a 13. Ferrandon, D., I. Koch, E. Westhof, and C. Nüsslein-Volhard. 1997.
RNA-RNA interaction is required for the formation of specific bicoid
stability element. We can find no strong similarities in either mRNA 3⬘ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16:
primary sequence or predicted secondary structure, but the 1751–1758.
uncertainties surrounding the nature of the BSF binding site 14. Fisk, D. G., M. B. Walker, and A. Barkan. 1999. Molecular cloning of the
maize gene crp1 reveals similarity between regulators of mitochondrial and
make it difficult to draw any conclusions about a later role for chloroplast gene expression. EMBO J. 18:2621–2630.
BSF in RNA stabilization during embryogenesis. Nevertheless, 15. Gunkel, N., T. Yano, F. H. Markussen, L. C. Olsen, and A. Ephrussi. 1998.
BSF is present throughout embryogenesis (data not shown), Localization-dependent translation requires a functional interaction be-
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guest
tween the 5⬘ and 3⬘ ends of oskar mRNA. Genes Dev. 12:1652–1664.
and thus its function is probably not limited to stabilization of 16. Henzel, W. J., T. M. Billeci, J. T. Stults, S. C. Wong, C. Grimley, and C.
maternal mRNAs, independent of which maternal mRNAs it Watanabe. 1993. Identifying proteins from two-dimensional gels by molec-
ular mass searching of peptide fragments in protein sequence databases.
regulates.
Proc. Natl. Acad. Sci. USA 90:5011–5015.
17. Kim-Ha, J., K. Kerr, and P. M. Macdonald. 1995. Translational regulation of
ACKNOWLEDGMENTS oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell
81:403–412.
This work was supported by grant GM42612 from the National 18. Krutzsch, H. C., and J. K. Inman. 1993. N-isopropyliodoacetamide in the
Institutes of Health to P.M.M. reduction and alkylation of proteins: use in microsequence analysis. Anal.
We thank members of the Macdonald lab for helpful discussions, Biochem. 209:109–116.
Karen Kerr and Yemane Geddes for technical assistance, Amy Beaton 19. Lai, E. C., C. Burks, and J. W. Posakony. 1998. The K box, a conserved 3⬘
of the BDGP for P element insertion stock l(2)k07109, the Blooming- UTR sequence motif, negatively regulates accumulation of enhancer of split
ton Stock Center for fly stocks, and Tim Stearns for monoclonal anti- complex transcripts. Development 125:4077–4088.
20. Lai, E. C., and J. W. Posakony. 1997. The Bearded box, a novel 3⬘ UTR
tubulin antibody. Mike Simon graciously provided lab space at a crit-
sequence motif, mediates negative post-transcriptional regulation of Bearded
ical juncture. and Enhancer of split complex gene expression. Development 124:4847–4856.
21. Lie, Y. S., and P. M. Macdonald. 1999. Apontic binds the translational
REFERENCES repressor Bruno and is implicated in regulation of oskar mRNA translation.
1. Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Development 126:1129–1138.
Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, 22. Lipshitz, H. D., and C. A. Smibert. 2000. Mechanisms of RNA localization
S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. and translational regulation. Curr. Opin. Genet. Dev. 10:476–488.
Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. 23. Lui, M., P. Tempst, and H. Erdjument-Bromage. 1996. Methodical analysis
Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. of protein-nitrocellulose interactions to design a refined digestion protocol.
Baxter, G. Helt, C. R. Nelson, G. L. Gabor Miklos, J. F. Abril, A. Agbayani, Anal. Biochem. 241:156–166.
H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. 24. Macdonald, P. M. 1990. bicoid mRNA localization signal: phylogenetic con-
Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. servation of function and RNA secondary structure. Development 110:161–
Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, 171.
P. Brokstein, P. Brottier, K. C. Burtis, D. A. Busam, H. Butler, E. Cadieu, A. 25. Macdonald, P. M., P. Ingham, and G. Struhl. 1986. Isolation, structure and
Center, I. Chandra, J. M. Cherry, S. Cawley, C. Dahlke, L. B. Davenport, P. expression of even-skipped: a second pair-rule gene of Drosophila containing
Davies, B. de Pablos, A. Delcher, Z. Deng, A. D. Mays, I. Dew, S. M. Dietz, a homeo box. Cell 47:721–734.
K. Dodson, L. E. Doup, M. Downes, S. Dugan-Rocha, B. C. Dunkov, P. 26. Macdonald, P. M., and K. Kerr. 1998. Mutational analysis of an RNA
Dunn, K. J. Durbin, C. C. Evangelista, C. Ferraz, S. Ferriera, W. Fleisch- recognition element that mediates localization of bicoid mRNA. Mol. Cell.
mann, C. Fosler, A. E. Gabrielian, N. S. Garg, W. M. Gelbart, K. Glasser, A. Biol. 18:3788–3795.
Glodek, F. Gong, J. H. Gorrell, Z. Gu, P. Guan, M. Harris, N. L. Harris, D. 27. Macdonald, P. M., and K. Kerr. 1997. Redundant RNA recognition events
Harvey, T. J. Heiman, J. R. Hernandez, J. Houck, D. Hostin, K. A. Houston, in bicoid mRNA localization. RNA 3:1413–1420.
T. J. Howland, M. H. Wei, C. Ibegwam, et al. 2000. The genome sequence of 28. Macdonald, P. M., K. Kerr, J. L. Smith, and A. Leask. 1993. RNA regulatory
Drosophila melanogaster. Science 287:2185–2195. element BLE1 directs the early steps of bicoid mRNA localization. Devel-
2. Arnott, D., W. J. Henzel, and J. T. Stults. 1998. Rapid identification of opment 118:1233–1243.
comigrating gel-isolated proteins by ion trap-mass spectrometry. Electro- 29. Macdonald, P. M., A. Leask, and K. Kerr. 1995. exl protein specifically binds
phoresis 19:968–980. BLE1, a bicoid mRNA localization element, and is required for one phase of
3. Arnott, D., K. L. O’Connell, K. L. King, and J. T. Stults. 1998. An integrated its activity. Proc. Natl. Acad. Sci. USA 92:10787–10791.
approach to proteome analysis: identification of proteins associated with 30. Macdonald, P. M., and G. Struhl. 1988. cis-acting sequences responsible for
cardiac hypertrophy. Anal. Biochem. 258:1–18. anterior localization of bicoid mRNA in Drosophila embryos. Nature 336:
4. Barkan, A., M. Walker, M. Nolasco, and D. Johnson. 1994. A nuclear 595–598.
mutation in maize blocks the processing and translation of several chloro- 31. Manthey, G. M., and J. E. McEwen. 1995. The product of the nuclear gene
plast mRNAs and provides evidence for the differential translation of alter- PET309 is required for translation of mature mRNA and stability or pro-
native mRNA forms. EMBO J. 13:3170–3181. duction of intron-containing RNAs derived from the mitochondrial COX1
5. Bashirullah, A., R. L. Cooperstock, and H. D. Lipshitz. 1998. RNA local- locus of Saccharomyces cerevisiae. EMBO J. 14:4031–4043.
ization in development. Annu. Rev. Biochem. 67:335–394. 32. Sallés, F. J., M. E. Lieberfarb, C. Wreden, J. P. Gergen, and S. Strickland.
6. Bashirullah, A., S. R. Halsell, R. L. Cooperstock, M. Kloc, A. Karaiskakis, 1994. Coordinate initiation of Drosophila development by regulated polyad-
W. W. Fisher, W. Fu, J. K. Hamilton, L. D. Etkin, and H. D. Lipshitz. 1999. enylation of maternal messenger RNAs. Science 266:1996–1999.
Joint action of two RNA degradation pathways controls the timing of ma- 33. Small, I. D., and N. Peeters. 2000. The PPR motif—a TPR-related motif
ternal transcript elimination at the midblastula transition in Drosophila prevalent in plant organellar proteins. Trends Biochem. Sci. 25:46–47.
melanogaster. EMBO J. 18:2610–2620. 34. Smibert, C. A., J. E. Wilson, K. Kerr, and P. M. Macdonald. 1996. smaug
7. Binder, R., J. A. Horowitz, J. P. Basilion, D. M. Koeller, R. D. Klausner, and protein represses translation of unlocalized nanos mRNA in the Drosophila
J. B. Harford. 1994. Evidence that the pathway of transferrin receptor embryo. Genes Dev. 10:2600–2609.
mRNA degradation involves an endonucleolytic cleavage within the 3⬘ UTR 35. Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty, N. Mozden, S.
and does not involve poly(A) tail shortening. EMBO J. 13:1969–1980. Misra, and G. M. Rubin. 1999. The Berkeley Drosophila Genome Project
8. Driever, W., and C. Nüsslein-Volhard. 1988. The bicoid protein determines gene disruption project: single P-element insertions mutating 25% of vital
position in the Drosophila embryo in a concentration-dependent manner. Drosophila genes. Genetics 153:135–177.
Cell 54:95–104. 36. Stephenson, E. C., Y. Chao, and J. D. Fackenthal. 1988. Molecular analysisVOL. 21, 2001 BSF STABILIZES bcd mRNA 3471
of the swallow gene of Drosophila melanogaster. Genes Dev. 2:1655–1665. stability elements in the ␣2-globin 3⬘ nontranslated region. Mol. Cell. Biol.
37. St. Johnston, D., W. Driever, T. Berleth, S. Richstein, and C. Nüsslein- 15:2457–2465.
Volhard. 1989. Multiple steps in the localization of bicoid RNA to the 40. Wickens, M., E. B. Goodwin, J. Kimble, S. Strickland, and M. Hentze. 2000.
anterior pole of the Drosophila oocyte. Development 107(Suppl.):13–19. Translational control of developmental decisions, p. 295–370. In N. Sonen-
38. Surdej, P., and M. Jacobs-Lorena. 1998. Developmental regulation of bicoid berg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene
mRNA stability is mediated by the first 43 nucleotides of the 3⬘ untranslated expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
region. Mol. Cell. Biol. 18:2892–2900. 41. Zinn, K., D. DiMaio, and T. Maniatis. 1983. Identification of two distinct
39. Weiss, I. M., and S. A. Liebhaber. 1995. Erythroid cell-specific mRNA regulatory regions adjacent to the human b-interferon gene. Cell 34:865–879.
Downloaded from http://mcb.asm.org/ on November 1, 2015 by guestYou can also read